Software:TPP
From SPCTools
| Revision as of 17:19, 1 June 2007 Jtasman (Talk | contribs) (→Source code (For Linux systems)) ← Previous diff |
Revision as of 17:20, 1 June 2007 Jtasman (Talk | contribs) (→Installing on Ubuntu) Next diff → |
||
| Line 17: | Line 17: | ||
| ====[[TPP:Installing_on_Ubuntu_7.04|Installing on Ubuntu]]==== | ====[[TPP:Installing_on_Ubuntu_7.04|Installing on Ubuntu]]==== | ||
| + | These community-contributed notes are a guide to installing on the Ubuntu distribution but should be similar to other linux systems. | ||
| ===[[TPP:Installing on Mac OSX|Mac OSX Installation guide]]=== | ===[[TPP:Installing on Mac OSX|Mac OSX Installation guide]]=== | ||
Revision as of 17:20, 1 June 2007
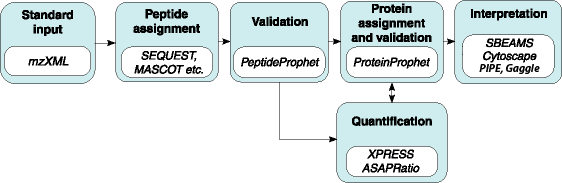
The Trans-Proteomic Pipeline (TPP) is a collection of integrated tools for MS/MS proteomics, developed at the SPC.
Contents |
Getting the software
Support Note
For support both during and after installation, you are highly recommended to consult the SPC Tools newsgroups:
- The spctools-discuss discussion group provides active community support and disucussion for the tools.
- Important information about new releases and software updates can be found on the spctools-announce discussion group. You are highly encouraged to subscibe to this low-volume list.
Installing on a Windows System
Source code Installation (For Linux systems)
The latest source code package can be found here, on the Sashimi project site on SourceForge.
Installing on Ubuntu
These community-contributed notes are a guide to installing on the Ubuntu distribution but should be similar to other linux systems.
Mac OSX Installation guide
Software contained in the TPP
Probability Assignment and Validation
PeptideProphet: Statistical validation of spectra-to-peptide sequence, using search engine results.
ProteinProphet: Protein identification and validation, using PeptideProphet results.
Protein Quantification
XPRESS: Calculation of relative abundance of proteins from MS/MS data.
ASAPRatio: Automated Statistical Analysis on Protein Ratio.
Libra: Four channel quantification software.
Graphical User Interface (GUI)
Petunia: Petunia is the name of the TPP's web-based GUI, which presents the tools in an organized and logical manner for those who do not wish to use the command-line.
Protein ID Curation
Out2Summary - converter of SEQUEST and TurboSEQUEST *.out files into a single HTML-SUMMARY file ready for use with INTERACT
Pep3D: Viewer for LC-MS and LC-MS/MS results.
Input Processing: mzXML Tools
readmzXML: mzXML parser based on RAMP
MsXML2Other: mzXML to SEQUEST dta, MASCOT generic and Micromass pkl converter
mzStar: SCIEX/ABI Analyst format to mzXML converter
ReAdW: ThermoFinnigan Xcalibur format to mzXML converter
RAMP: mzXML data parser
Input Processing: Search-Engine to pepXML converters
- Sequest results: Out2XML
- Mascot results: Mascot2XML
- Tandem results: Tandem2XML