Software:TPP
From SPCTools
| Revision as of 18:27, 11 April 2007 Jtasman (Talk | contribs) (adding link to readmzXML) ← Previous diff |
Revision as of 21:00, 13 April 2007 Jtasman (Talk | contribs) (adding links to spctools newsgroups) Next diff → |
||
| Line 5: | Line 5: | ||
| ==Getting the software== | ==Getting the software== | ||
| + | |||
| + | ===Support Note=== | ||
| + | For support both during and after installation, you are highly recommended to consult the SPC Tools newsgroups: | ||
| + | * The [http://groups.google.com/group/spctools-discuss spctools-discuss discussion group] provides active community support and disucussion for the tools. | ||
| + | * Important information about new releases and software updates can be found on the [http://groups.google.com/group/spctools-announce spctools-announce discussion group]. You are '''highly encouraged''' to subscibe to this low-volume list. | ||
| ===[[TPP:Windows Cygwin Installation|Installing on a Windows System]]=== | ===[[TPP:Windows Cygwin Installation|Installing on a Windows System]]=== | ||
| Line 19: | Line 24: | ||
| <!-- <P>Download source from Web: <a href="http://sourceforge.net/project/showfiles.php?group_id=69281">Sourceforge Page</a><br> --> | <!-- <P>Download source from Web: <a href="http://sourceforge.net/project/showfiles.php?group_id=69281">Sourceforge Page</a><br> --> | ||
| <!-- <em>Note: On the web you might find a newer version of this software.</em></P> --> | <!-- <em>Note: On the web you might find a newer version of this software.</em></P> --> | ||
| - | |||
| ==Software contained in the TPP== | ==Software contained in the TPP== | ||
Revision as of 21:00, 13 April 2007
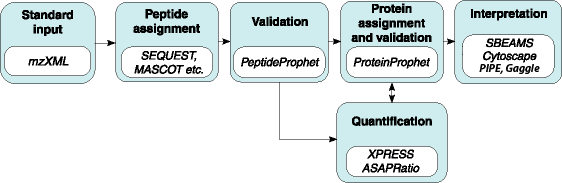
The Trans-Proteomic Pipeline (TPP) is a collection of integrated tools for MS/MS proteomics, developed at the SPC.
Contents |
Getting the software
Support Note
For support both during and after installation, you are highly recommended to consult the SPC Tools newsgroups:
- The spctools-discuss discussion group provides active community support and disucussion for the tools.
- Important information about new releases and software updates can be found on the spctools-announce discussion group. You are highly encouraged to subscibe to this low-volume list.
Installing on a Windows System
Source code (For Linux systems)
The latest source code package can be found here, on the Sashimi project site on SourceForge.
Mac OSX Installation guide
Software contained in the TPP
Probability Assignment and Validation
PeptideProphet: Statistical validation of spectra-to-peptide sequence, using search engine results.
ProteinProphet: Protein identification and validation, using PeptideProphet results.
Protein Quantification
XPRESS: Calculation of relative abundance of proteins from MS/MS data.
ASAPRatio: Automated Statistical Analysis on Protein Ratio.
Libra: Four channel quantification software.
Graphical User Interface (GUI)
Petunia: Petunia is the name of the TPP's web-based GUI, which presents the tools in an organized and logical manner for those who do not wish to use the command-line.
Protein ID Curation
Out2Summary - converter of SEQUEST and TurboSEQUEST *.out files into a single HTML-SUMMARY file ready for use with INTERACT
Pep3D: Viewer for LC-MS and LC-MS/MS results.
Input Processing: mzXML Tools
readmzXML: mzXML parser based on RAMP
MsXML2Other: mzXML to SEQUEST dta, MASCOT generic and Micromass pkl converter
mzStar: SCIEX/ABI Analyst format to mzXML converter
ReAdW: ThermoFinnigan Xcalibur format to mzXML converter
RAMP: mzXML data parser