Software:TIQAM
From SPCTools
| Revision as of 23:00, 18 April 2008 Vinz (Talk | contribs) (→Software Information) ← Previous diff |
Revision as of 23:12, 18 April 2008 Vinz (Talk | contribs) (→Installation) Next diff → |
||
| Line 18: | Line 18: | ||
| ===Step 2 - Create Databases for TIQAM software=== | ===Step 2 - Create Databases for TIQAM software=== | ||
| - | =====Option 1===== | + | |
| Please carefully type the following statments exactly as written | Please carefully type the following statments exactly as written | ||
| C:\where you installed mysql\bin \-user=root mysql \-p | C:\where you installed mysql\bin \-user=root mysql \-p | ||
| Line 28: | Line 28: | ||
| mysql> FLUSH PRIVILEGES; | mysql> FLUSH PRIVILEGES; | ||
| mysql> quit; | mysql> quit; | ||
| - | |||
| - | =====Option 2===== | ||
| - | *Download [[initialize.sql]] and put it in the folder where you installed MySQL. | ||
| - | *Go to the command line, navigate to the folder where you installed MySQL. | ||
| - | *Type: mysql \-u root \-p < initialize.sql\\ | ||
| ===Step 3 - Initialize the TIQAM Digestor database=== | ===Step 3 - Initialize the TIQAM Digestor database=== | ||
Revision as of 23:12, 18 April 2008
Contents |
Introduction
TIQAM (Targeted Identification for Quantitative Analysis by MRM) is developed as a suite of software tools to support targeted indentification and quantification using multiple reaction monitoring (MRM) mass spectum technology. More specifically, TIQAM software provided user friendly interfaces to assist process of peptide selection, transition generation and validation. The overview of the software supporting work flow is illustrated by the following figure.
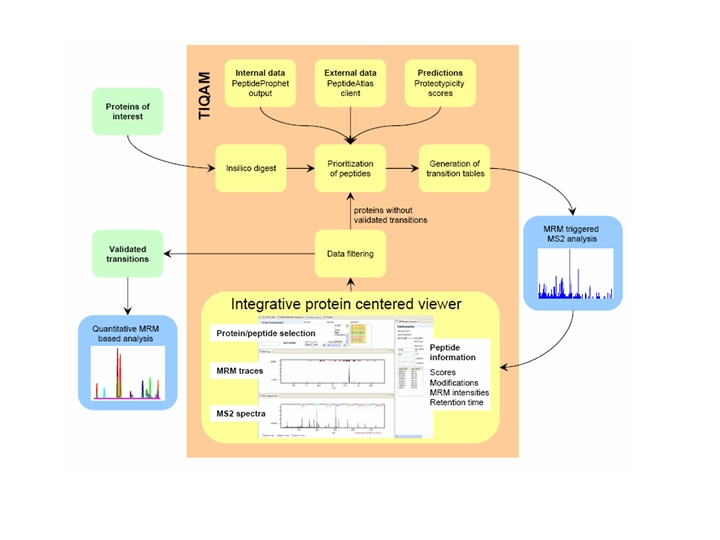
Detailed information and reference
The detailed information on the work flow has been published: Lange V, Malmstrom JA, Didion J, King NL, Johansson BP, Schafer J, Rameseder J, Wong C-H, Deutsch EW, Brusniak M-Y, Buhlmann P, Bjorck L, Domon B, Aebersold R (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics: M800032-MCP800200
Installation
Current TIQAM software supports Window, Mac and Linux OS and requires Java 1.6 java run time environment and MySQL community edition 5.0 database installed prior to running TIQAM software. TIQAM software can be downloaded from the TIQAM download site.
Step 1 - Install MySQL
Prior to starting TIQAM install MySQL
- Download MySQL server from mysql site.
- Installation instructions are available on the mysql.com site. For example, the installation instructions for Windows are available at mysql documentation site.
- Important: Select transactional database (InnoDB)!
- Choose a drive where you have sufficient diskspace for the database (several gb).
- Start MySQL prior to running any of TIQAM software. The automatic start-up of your installed database server can be found at mysql document site.
Step 2 - Create Databases for TIQAM software
Please carefully type the following statments exactly as written
C:\where you installed mysql\bin \-user=root mysql \-p mysql> create database trans; mysql> create database mrm; mysql> use mysql; mysql> grant all privileges on trans.\* to 'm'@'localhost' identified by 'mmm' with grant option; mysql> grant all privileges on mrm.\* to 'm'@'localhost' identified by 'mmm' with grant option; mysql> FLUSH PRIVILEGES; mysql> quit;
Step 3 - Initialize the TIQAM Digestor database
Start TIQAM Digestor. You will see an error message saying "Cannot execute query." Click OK. In the left-hand navigation panel, click the right-arrow to expand the navigator. Double-click "Database Helper." Now click "Test Drop DB then Create DB."
Step 4 - Download TIQAM and unzip
Unzip the downloaded files. Make sure, you keep the folder structure when extracting! You will have 3 folders with TIQAM-digester, TIQAM-peptide-atlas and TIQAM-viewer (with version number suffixes). In each folder subdirectory, you will find .exe files to launch the TIQAM module.
Contact Information
Mi-Youn Brusniak, Ph.D. 1441 North 34th St. Seattle, WA 98103 USA mbrusniak@systemsbiology.org

