TIQAM (Targeted Identification for Quantitative Analysis by MRM)
Introduction
TIQAM is developed as a suite of software tools to support targeted indentification and quantification using multiple reaction monitoring (MRM) mass spectum technology. More specifically, TIQAM software provided user friendly interfaces to assist process of peptide selection, transition generation and validation. The overview of the software supporting work flow is illustrated by the following figure.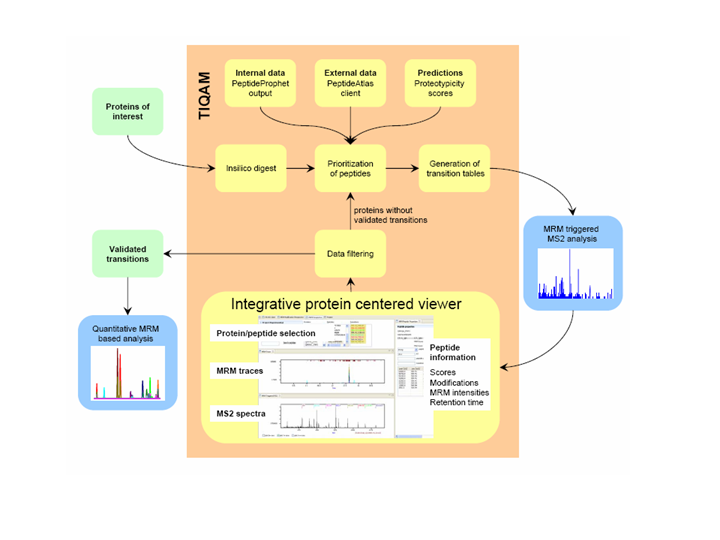
The detailed information on the work flow and the application using TIQAM software for targeted quantitative analysis of Streptococcus pyogenes virulenece factors has been written and submitted to a scientific journal for review. The article title and authors are "Targeted Quantitative Analaysis of Streptococcus Pyogenes Virulenece Factors by Multiple Reaction Monitoring" by
Vinzenz
Lange, Johan A. Malmström,
Third party software requirement
Current TIQAM software supports Window, Mac and Linux OS and requires Java 1.6 java run time environment and MySQL community edition 5.0 database installed prior to running TIQAM software.Installing MySQL
- download MySQL server, by unzipping mysql.zip and follow mysql instruction.
- installation instructions are available in mysql.com site. For example, the installation instructions for Windows are available at mysql document site.
- Start MySQL prior to running any of TIQAM software. The automatic start-up of your installed database server can be found at mysql document site.
Create Databases for TIQAM software
Please download sql scripts (createtrans.sql, createmrm.sql) carefully type the following statments exactly as written>C:\where you installed mysql\bin\mysql -u root -p < createtrans.sql
>C:\where you installed mysql\bin\mysql -u root -p < createmrm.sql
Download and installing mzWiff for wiff file converter to mzXML
TIQAM software uses mzXML as an input standard. The mzWiff is a wiff file converter to mzXML.mzWiff program can be downloaded from mzWiff beta version website.
Current release version of TIQAM software suite
TIQAM-Digestor 1.2.3TIQAM-PeptideAtlas 1.0.3
TIQAM-Viewer 1.2.2
Download and installing TIQAM software suite
TIQAM for Win32_x86
TIQAM for Linux_gtk_x86
Unzip the download folder and you can find TIQAM-digester, TIQAM-peptide-atlas and TIQAM-viewer with version number suffix folders. In each folder subdirectory, you will find .exe files to launch each TIQAM modules.
The corresponding demo data set is in TIQAM-TutorialDemoData.zip
If you have any question on software installation, please contact using the following contact information.
Copyright (c) 2007 Mi-Youn Brusniak and Aebersold groupTIQAM for Linux_gtk_x86
Unzip the download folder and you can find TIQAM-digester, TIQAM-peptide-atlas and TIQAM-viewer with version number suffix folders. In each folder subdirectory, you will find .exe files to launch each TIQAM modules.
Download Tutorial Documentations and Demo Data Set
Tutorial documents for each module is in TIQAM-Tutorial.zipThe corresponding demo data set is in TIQAM-TutorialDemoData.zip
Download Data Set related to the above reference paper published in Molecular & Cellular Proteomics 2008
DataSet for TIQAMDownload TIQAM source code
TIQAM-src.tgzMore information is available in the following wiki site.
TIQAM Wiki at Seattle Proteomics CenterIf you have any question on software installation, please contact using the following contact information.
Contact information:
Mi-Youn Brusniak, Ph.D.
1441 North 34th St.
Seattle, WA 98103 USA
mbrusniak@systemsbiology.org