Software:TIQAM
From SPCTools
| Revision as of 22:40, 4 April 2008 Mbrusniak (Talk | contribs) ← Previous diff |
Revision as of 22:23, 18 April 2008 Vinz (Talk | contribs) (→Detailed information and reference) Next diff → |
||
| Line 3: | Line 3: | ||
| [[Image:TIQAMWorkFlow.jpg]] | [[Image:TIQAMWorkFlow.jpg]] | ||
| ==Detailed information and reference== | ==Detailed information and reference== | ||
| - | The detailed information on the work flow and the application using TIQAM software for targeted quantitative analysis of Streptococcus pyogenes virulenece factors has been written and submitted to a scientific journal for review. The article title and authors are "Targeted Quantitative Analaysis of Streptococcus Pyogenes Virulenece Factors by Multiple Reaction Monitoring" by | + | The detailed information on the work flow has been published: |
| - | + | [http://www.mcponline.org/cgi/content/abstract/M800032-MCP200v1|Lange V, Malmstrom JA, Didion J, King NL, Johansson BP, Schafer J, Rameseder J, Wong C-H, Deutsch EW, Brusniak M-Y, Buhlmann P, Bjorck L, Domon B, Aebersold R (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics: M800032-MCP800200] | |
| - | Vinzenz Lange, Johan A. Malmström, John Didion, Nichole L. King, Björn P. Johansson, Juliane Schäfer, Jonathan Rameseder, Chee-Hong Wong, Eric W. Deutsch, Mi-Youn Brusniak, Peter Bühlman, Lars Björck, Bruno Domon and Ruedi Aebersold. | + | |
| ==Software Information== | ==Software Information== | ||
Revision as of 22:23, 18 April 2008
Contents |
Introduction
TIQAM (Targeted Identification for Quantitative Analysis by MRM) is developed as a suite of software tools to support targeted indentification and quantification using multiple reaction monitoring (MRM) mass spectum technology. More specifically, TIQAM software provided user friendly interfaces to assist process of peptide selection, transition generation and validation. The overview of the software supporting work flow is illustrated by the following figure.
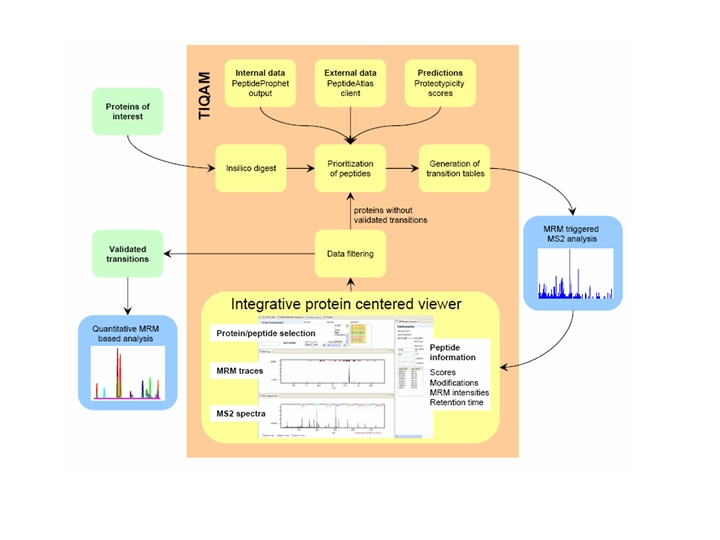
Detailed information and reference
The detailed information on the work flow has been published: V, Malmstrom JA, Didion J, King NL, Johansson BP, Schafer J, Rameseder J, Wong C-H, Deutsch EW, Brusniak M-Y, Buhlmann P, Bjorck L, Domon B, Aebersold R (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics: M800032-MCP800200
Software Information
Current TIQAM software supports Window, Mac and Linux OS and requires Java 1.6 java run time environment and MySQL community edition 5.0 database installed prior to running TIQAM software. TIQAM software can be download from the TIQAM download site.
Contact Information
Mi-Youn Brusniak, Ph.D. 1441 North 34th St. Seattle, WA 98103 USA mbrusniak@systemsbiology.org

