Software:TIQAM
From SPCTools
| Revision as of 23:32, 18 April 2008 Vinz (Talk | contribs) (→Detailed information and reference) ← Previous diff |
Current revision Vinz (Talk | contribs) (→Limitations) |
||
| Line 1: | Line 1: | ||
| ==Introduction== | ==Introduction== | ||
| - | TIQAM (Targeted Identification for Quantitative Analysis by MRM) is developed as a suite of software tools to support targeted indentification and quantification using multiple reaction monitoring (MRM) mass spectum technology. More specifically, TIQAM software provided user friendly interfaces to assist process of peptide selection, transition generation and validation. The overview of the software supporting work flow is illustrated by the following figure. | + | TIQAM (Targeted Identification for Quantitative Analysis by MRM) is developed as a suite of software tools to support targeted indentification and quantification using multiple reaction monitoring (MRM) mass spectum technology. More specifically, TIQAM software provided user friendly interfaces to assist process of peptide selection, transition generation and validation. The overview of the software supporting work flow is illustrated by the following figure ([http://www.mcponline.org/cgi/content/abstract/M800032-MCP200v1 Lange et al. 2008 Mol Cell Proteomics]). |
| + | |||
| [[Image:TIQAMWorkFlow.jpg]] | [[Image:TIQAMWorkFlow.jpg]] | ||
| + | |||
| ==Detailed information and reference== | ==Detailed information and reference== | ||
| TIQAM currently consists of 3 modules: | TIQAM currently consists of 3 modules: | ||
| Line 12: | Line 14: | ||
| All data and user input is saved in the mySQL database. So you may stop and restart at any time. However, reports (exports) are not saved in the database. | All data and user input is saved in the mySQL database. So you may stop and restart at any time. However, reports (exports) are not saved in the database. | ||
| ====Import of data==== | ====Import of data==== | ||
| + | Before importing data make sure, all modifications are correctly defined. The names of the modifications in the csv file and on the modification page need to match. | ||
| TIQAM expects the following data formats: | TIQAM expects the following data formats: | ||
| * Centroided [[Formats:mzXML|mzXML]] data (use [http://web.bii.a-star.edu.sg/~wongch/mzWiff/ mzWiff] for conversion of data from ABI/MDS Sciex instruments.) | * Centroided [[Formats:mzXML|mzXML]] data (use [http://web.bii.a-star.edu.sg/~wongch/mzWiff/ mzWiff] for conversion of data from ABI/MDS Sciex instruments.) | ||
| * A csv file with the list of transitions and associated proteins. Such a file is generated by TIQAM-digestor. | * A csv file with the list of transitions and associated proteins. Such a file is generated by TIQAM-digestor. | ||
| - | * pepXML files. This file is generated by the TPP when xinteract is run. | + | * pepXML files. This file is generated by the [[Software:TPP|TPP]] when xinteract is run. |
| ====Basic operation==== | ====Basic operation==== | ||
| - | Start by selecting a project and experiment. Then select proteins and peptides to view. In the MRM trace window you will see the traces. The tick marks at the top indicate the aquisition of MS2 spectra. Clicking in the trace window will update the MS2 spectrum. The current MS2 spectrum is indicated by a red arrow in the MRM-trace window. You may zoom by drawing a rectangle with the mouse. Right click unzooms again. | + | Start by selecting a project and experiment. Then select proteins and peptides to view. In the MRM trace window you will see the traces. The tick marks at the top indicate the acquisition of MS2 spectra. Clicking in the trace window will update the MS2 spectrum. The current MS2 spectrum is indicated by a red arrow in the MRM-trace window. You may zoom by drawing a rectangle with the mouse. Right click unzooms again. |
| MS2 spectra have the matching B, Y or Y+\+ ions colored in the color of the series. If you click into the MS2 spectrum, you can scroll to the next acquired MS2 spectrum by the right or left arrow key. | MS2 spectra have the matching B, Y or Y+\+ ions colored in the color of the series. If you click into the MS2 spectrum, you can scroll to the next acquired MS2 spectrum by the right or left arrow key. | ||
| Line 29: | Line 32: | ||
| ====Exporting transitions==== | ====Exporting transitions==== | ||
| - | Select the project and experiment you want to export transitions. Make your selections. You will get an overview of proteins, peptides and the transitions. If you are not fully happy with the list and want to change your choice, press the yellow arrow. You may then create a new report with modified paramaters. | + | Select the project and experiment you want to export transitions. Make your selections. You will get an overview of proteins, peptides and the transitions. If you are not fully happy with the list and want to change your choice, press the yellow arrow. You may then create a new report with modified parameters. |
| ===TIQAM-Peptide Atlas Client=== | ===TIQAM-Peptide Atlas Client=== | ||
| Line 51: | Line 54: | ||
| ===Step 2 - Create Databases for TIQAM software=== | ===Step 2 - Create Databases for TIQAM software=== | ||
| - | + | ====Option 1==== | |
| Please carefully type the following statments exactly as written | Please carefully type the following statments exactly as written | ||
| C:\where you installed mysql\bin \-user=root mysql \-p | C:\where you installed mysql\bin \-user=root mysql \-p | ||
| Line 61: | Line 64: | ||
| mysql> FLUSH PRIVILEGES; | mysql> FLUSH PRIVILEGES; | ||
| mysql> quit; | mysql> quit; | ||
| + | ====Option 2==== | ||
| + | Download the file [http://groups.google.com/group/tiqam/web/initialize.sql initialize] and put it in the folder where you installed mySQL. | ||
| - | ===Step 3 - Initialize the TIQAM Digestor database=== | + | Go to the command line, navigate to the folder where you installed mySQL. |
| - | Start TIQAM Digestor. You will see an error message saying "Cannot execute query." Click OK. In the left-hand navigation panel, click the right-arrow to expand the navigator. Double-click "Database Helper." Now click "Test Drop DB then Create DB." | + | |
| - | ===Step 4 - Download TIQAM and unzip=== | + | Type: mysql -u root -p < initialize.sql |
| + | |||
| + | |||
| + | |||
| + | ===Step 3 - Download TIQAM and unzip=== | ||
| Unzip the [http://tools.proteomecenter.org/TIQAM/TIQAM.html downloaded files]. Make sure, you keep the folder structure when extracting! You will have 3 folders with TIQAM-digester, TIQAM-peptide-atlas and TIQAM-viewer (with version number suffixes). In each folder subdirectory, you will find .exe files to launch the TIQAM module. | Unzip the [http://tools.proteomecenter.org/TIQAM/TIQAM.html downloaded files]. Make sure, you keep the folder structure when extracting! You will have 3 folders with TIQAM-digester, TIQAM-peptide-atlas and TIQAM-viewer (with version number suffixes). In each folder subdirectory, you will find .exe files to launch the TIQAM module. | ||
| + | |||
| + | ==Current Limitations== | ||
| + | TIQAM was developed to '''validate and optimize''' transitions for MRM measurements. | ||
| + | * It is not a quantification software. In particular it does not feature the quantitative comparison of several sample runs. Currently all transitions measured for a particular peptide and imported into the same project will be displayed together. | ||
| + | * It does not include a database search engine. Searches have to be performed before importing data into TIQAM-viewer. However, you may view your data without search scores if you did not search them. | ||
| + | |||
| + | ===Limitations=== | ||
| + | * TIQAM was only tested with data acquired on a Q-TRAP 4000, converted with [[Software:mzWiff|mzWIFF]]. If you intend to use TIQAM for data from other instruments please [http://groups.google.com/group/tiqam let us know] and we will work with you to get the formats compatible. | ||
| + | |||
| + | ===Feedback and Questions=== | ||
| + | Please join the [http://groups.google.com/group/tiqam TIQAM discussion group] for feature requests, questions and bug reports. | ||
| ==Contact Information== | ==Contact Information== | ||
Current revision
Contents |
Introduction
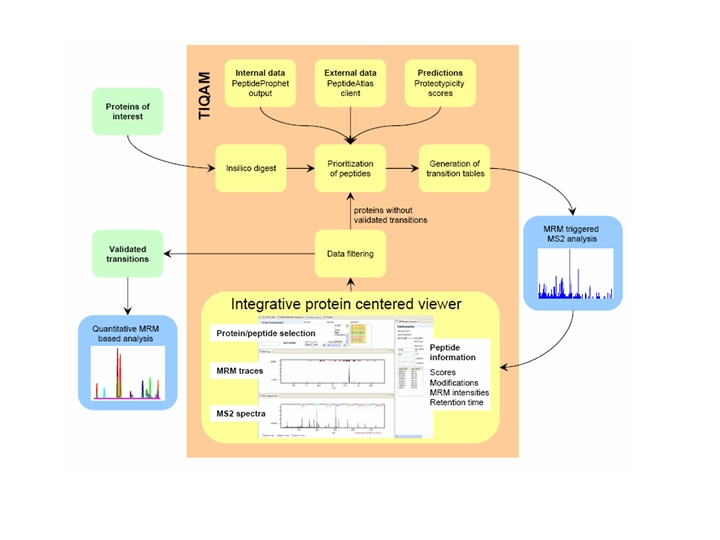
TIQAM (Targeted Identification for Quantitative Analysis by MRM) is developed as a suite of software tools to support targeted indentification and quantification using multiple reaction monitoring (MRM) mass spectum technology. More specifically, TIQAM software provided user friendly interfaces to assist process of peptide selection, transition generation and validation. The overview of the software supporting work flow is illustrated by the following figure (Lange et al. 2008 Mol Cell Proteomics).
Detailed information and reference
TIQAM currently consists of 3 modules:
TIQAM-digestor
TIQAM-digestor is used to create a list of transitions for MRM measurements. Starting from a fasta file with proteins, peptides are generated by digestion (with options to include and exclude peptides based on pattern matching). To prioritize peptides which should be targeted, pepXML files or tables (format: peptide, number) can be uploaded. Peptides are sorted within the proteins. Transitions are generated based on user criteria.
TIQAM-viewer
In TIQAM-viewer MRM and MRM triggered MS2 data can be viewed to validate transitions. Best transitions can be exported for quantification. All data and user input is saved in the mySQL database. So you may stop and restart at any time. However, reports (exports) are not saved in the database.
Import of data
Before importing data make sure, all modifications are correctly defined. The names of the modifications in the csv file and on the modification page need to match.
TIQAM expects the following data formats:
- Centroided mzXML data (use mzWiff for conversion of data from ABI/MDS Sciex instruments.)
- A csv file with the list of transitions and associated proteins. Such a file is generated by TIQAM-digestor.
- pepXML files. This file is generated by the TPP when xinteract is run.
Basic operation
Start by selecting a project and experiment. Then select proteins and peptides to view. In the MRM trace window you will see the traces. The tick marks at the top indicate the acquisition of MS2 spectra. Clicking in the trace window will update the MS2 spectrum. The current MS2 spectrum is indicated by a red arrow in the MRM-trace window. You may zoom by drawing a rectangle with the mouse. Right click unzooms again. MS2 spectra have the matching B, Y or Y+\+ ions colored in the color of the series. If you click into the MS2 spectrum, you can scroll to the next acquired MS2 spectrum by the right or left arrow key.
Validating transitions
To validate a transition select the MS2 spectra at the MRM-trace maximum. Choose a peptide validation human score and press save. The peak height of the transitions at that time will be saved. If no MS2 spectrum was acquired at the peak maximum you may manually type the correct retention time and press save.
Exporting transitions
Select the project and experiment you want to export transitions. Make your selections. You will get an overview of proteins, peptides and the transitions. If you are not fully happy with the list and want to change your choice, press the yellow arrow. You may then create a new report with modified parameters.
TIQAM-Peptide Atlas Client
The Peptide Atlas Client connects to the PeptideAtlas to retrieve information about PTPs (Proteotypic Peptides).
Reference
Detailed information on the work flow has been published:
Installation
Current TIQAM software supports Window, Mac and Linux OS and requires Java 1.6 java run time environment and MySQL community edition 5.0 database installed prior to running TIQAM software. TIQAM software can be downloaded from the TIQAM download site.
Step 1 - Install MySQL
Prior to starting TIQAM install MySQL
- Download MySQL server from mysql site.
- Installation instructions are available on the mysql.com site. For example, the installation instructions for Windows are available at mysql documentation site.
- Important: Select transactional database (InnoDB)!
- Choose a drive where you have sufficient diskspace for the database (several gb).
- Start MySQL prior to running any of TIQAM software. The automatic start-up of your installed database server can be found at mysql document site.
Step 2 - Create Databases for TIQAM software
Option 1
Please carefully type the following statments exactly as written
C:\where you installed mysql\bin \-user=root mysql \-p mysql> create database trans; mysql> create database mrm; mysql> use mysql; mysql> grant all privileges on trans.\* to 'm'@'localhost' identified by 'mmm' with grant option; mysql> grant all privileges on mrm.\* to 'm'@'localhost' identified by 'mmm' with grant option; mysql> FLUSH PRIVILEGES; mysql> quit;
Option 2
Download the file initialize and put it in the folder where you installed mySQL.
Go to the command line, navigate to the folder where you installed mySQL.
Type: mysql -u root -p < initialize.sql
Step 3 - Download TIQAM and unzip
Unzip the downloaded files. Make sure, you keep the folder structure when extracting! You will have 3 folders with TIQAM-digester, TIQAM-peptide-atlas and TIQAM-viewer (with version number suffixes). In each folder subdirectory, you will find .exe files to launch the TIQAM module.
Current Limitations
TIQAM was developed to validate and optimize transitions for MRM measurements.
- It is not a quantification software. In particular it does not feature the quantitative comparison of several sample runs. Currently all transitions measured for a particular peptide and imported into the same project will be displayed together.
- It does not include a database search engine. Searches have to be performed before importing data into TIQAM-viewer. However, you may view your data without search scores if you did not search them.
Limitations
- TIQAM was only tested with data acquired on a Q-TRAP 4000, converted with mzWIFF. If you intend to use TIQAM for data from other instruments please let us know and we will work with you to get the formats compatible.
Feedback and Questions
Please join the TIQAM discussion group for feature requests, questions and bug reports.
Contact Information
Mi-Youn Brusniak, Ph.D. 1441 North 34th St. Seattle, WA 98103 USA mbrusniak@systemsbiology.org