
Seattle Proteome Center (SPC) - Proteomics Tools
NHLBI Proteomics Center at the Institute for Systems Biology
About the Center:
|
|
|
|
XLink Package- Version 0.93 online manual -- Version 0.92 online manual- A new set of computer analysis tools for MALDI mass spec data derived from digested samples of crosslinked protein complexes. The software used for automated analysis contains three programs: iXLINK, a PERL program executed from the command line, doXLINK, a library of java programs executed by a PERL script which is also executed from the command line, and the java program XLinkViewer. 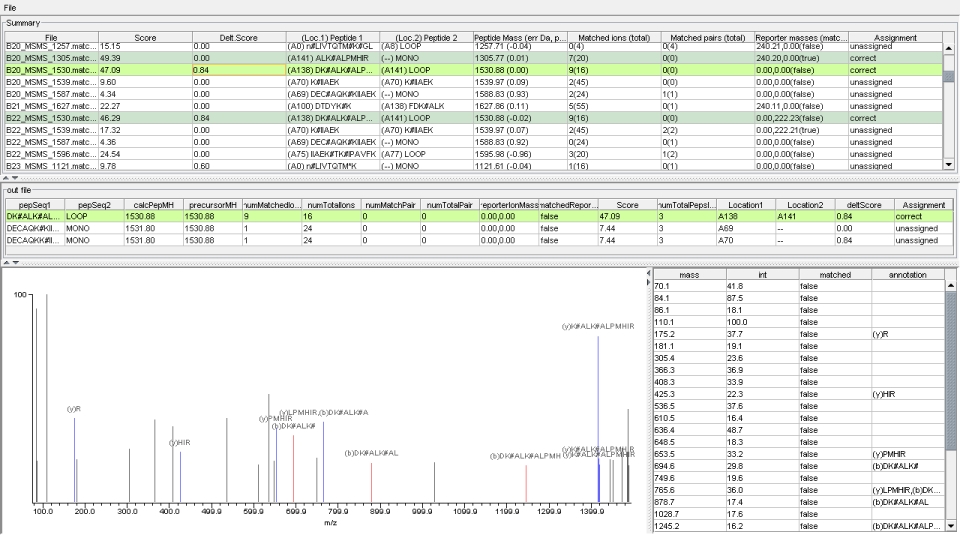 Method outline: Prior to computational analysis, two crosslinked peptide mixtures are independently fractionated by reverse phase column chromatography and spotted onto two standard MALDI plates. Each mixture contains peptides modified with both the isotopically heavy and light version of a crosslinking reagent. Consequently, modified peptides should exist within a given spectrum as a doublet - a pair of peaks separated by the mass difference between the heavy and light form of the label. One of the samples is reacted with crosslinking reagent in buffer containing [16O] water. The other is reacted in buffer containing a mix of [16O] and [18O] water. Within the [16O] / [18O] mixture, monolink peaks will appear as quadruplets whereas crosslink peaks will appear as doublets. This method can also be used without [18O] water. Further details of the experimental procedure are given in the original publication [1]. iXLINK classifies peaks from a duplex LC-MALDI experiment into one of four categories: noise, high abundance, crosslink-derived and monolink-derived. iXLINK generates a database of crosslinker-modified species based on protein sequence(s), crosslinker and protease(s). Once classified, mass mapping is used to generate a preliminary sequence assignment. iXLINK also creates mass inclusion lists for subsequent MS/MS acquisition of potentially crosslinker-modified peptide species. doXLINK provides matching scores for MS/MS data, that was acquired based on mass inclusionlists created by iXLINK, to peptides included in iXLINK's preliminary sequence assignment. XLinkViewer is used to visualize iXLINK and doXLINK results, and allows the user to confirm or reject the doXLINK assignments. [1] Seebacher, J., Mallick, P., Zhang, N., Eddes, J. S., Aebersold, R., Gelb, M. H. (2006) "Integrative Protein Crosslinking Analysis Using Mass Spectrometry, Isotope-Coded Crosslinkers, and Computational Data Processing" J. Proteome Res., Web Release Date: 22-Jul-2006 Version 0.93: Current version: XLINK_Software_Tools.zip Version 0.92: Old version: XLINK_Software_Tools.zip |
