Corra
Corra is a single, user-friendly, informatic framework, that is simple to use and fully customizable, for the enabling of LC-MS-based quantitative proteomic workflows of any size, able to guide the user seamlessly from MS data generation, through data processing, visualization, and statistical analysis steps, to the identification of differentially abundant or expressed candidate features for prioritized targeted identification by subsequent MS/MS.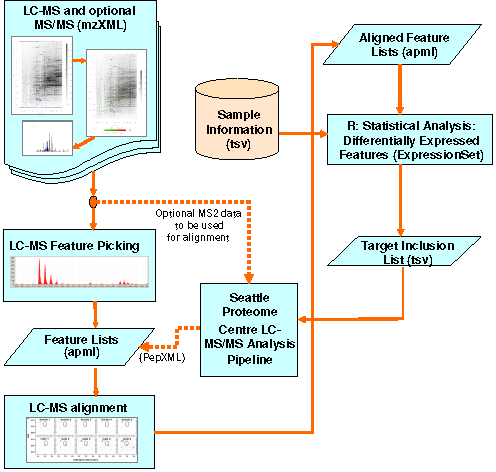
A goal of Corra was to enable the integration of multiple and disparate LC-MS data analysis tools, and integrate them, seamlessly, with common statistical packages to allow for better comparison between differently-processed datasets, via the addition of statistical measures of confidence and error rates. The integration of tools was achieve via
Corra and its biological application is published in the BMC
Bioinformatics 2008 9:542 and be accessible by http://www.biomedcentral.com/1471-2105/9/542
Corra and APML download site:
You can download source codes and documentation by http://sourceforge.net/projects/corra
You can download ftp://ftp.systemsbiology.org/Corra/example_mzXML.tgz file contains 6 mzXML files (1.3 GBtye) acquired from ThermoFinnigan LTQ Orbitrap data from yeast wild type and gene knockout dataset, which is described in the above paper.
Corra tutorial guides:
Corra v3.0 tutorial guide can be downloaded by CorraUserGuide-Tutorial.pdf
Corra-ATAQS developer code base and discussion group
Please join as project member for future development and biomedical research collaborators and please join for discussion.
SRM based hypothesis-driven computational tool
As a complementary to Corra tools, we recently developed and published hypothesis-driven computational tool called ATAQS.
Messages:
Thank you for
interested in using Corra for your biological data application or enhancing
current software or algorithm research.
We are looking
forward to hearing from you and future collaboration as a community based code
development.
Sincerely,
/**
* Copyright (c) 2008 Institute for Systems Biology
*
* Licensed under the Apache License, Version 2.0 (the "License");
* you may not use this file except in compliance with the License.
* You may obtain a copy of the License at
*
* http://www.apache.org/licenses/LICENSE-2.0
*
* Unless required by applicable law or agreed to in writing, software
* distributed under the License is distributed on an "AS IS" BASIS,
* WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
* See the License for the specific language governing permissions and
* limitations under the License.
*
* Contact info:
* Mi-Youn K. Brusniak
* Insitute for Systems Biology
* 1441 North 34th St.
* Seattle, WA 98103 USA
* mbrusniak@systemsbiology.org
*
*/
