ATAQS (Automated and
Targeted Analysis with Quantitative SRM)
As a complement to
the well-established discovery proteomic methods, targeted mass spectrometry
based on SRM is becoming an important tool for the generation of reproducible,
sensitive and quantitatively accurate data from biological samples. The method depends
on the generation of target protein sets based on prior information and the
one-time generation of verified mass spectrometric assays for each of the
targeted proteins. The development of these assays depends on the optimal
selection of peptides that represent the proteins on the target list and the
optimal set of transitions for their detection in biological samples. Once
developed, these assays can be continually applied across a multitude of
studies.
The ATAQS pipeline
(see figure below) and software provides a high throughput tool for organizing,
generating and verifying transition lists and for the post acquisition analysis
and dissemination of the data generated from applying the transition lists to
studies of biological samples. ATAQS uses information from publicly
accessible databases for the optimization of the protein and peptide target
lists and for the optimization of a transition set. ATAQS is an open source
software that enables data-driven researchers to generate candidate protein
lists and measure candidate proteins across a large number of biological
samples, and allows algorithm-developing scientists to further develop the
steps in the ATAQS pipeline. As needs arise, we plan to continuously expand on
ATAQS functionalities (e.g., validation of quantification, support of SILAC
type experiments, etc.)
The novel aspects of ATAQS are:
- Open source software for supporting large
scale SRM verification workflow by multi users or research sites by managing
projects and by running in a multi-threaded and distributed computing environment,
to enable data processing on a compute cluster. This allows for timely processing
of the typically large data sets generated in a high-throughput proteomics environment,
and also ensures that the client computer’s resources are not drained.
- ATAQS provides a user friendly interface to
access publically available transition resources to download and also
contribute user optimized transitions to public database if researchers choose
to do so.
- ATAQS produces an automated way of generating
all of your verified transitions in Proteomics Standard Initiative format for
SRM called TraML. Thus, it will be easier for exchanging data among proteomics
community.
- A
web-based GUI to provide the user with the ability to manage multiple projects
and data analyses, as well as direct the data through the analytical and
statistical modules, all via a web browser. The GUI also provides
significant user control to customize the process for
their needs and to output results in visual format as well as to generate TraML format
of measured transitions.
- ATAQS includes extensive documentation to
enable a bioinformatician to customize the ATAQS platform for their own and lab
uses, by implementation of additional analytical tools, Bioconductor
packages etc. as needed.
We expect that
ATAQS will find wide application as targeted proteomics increases in use to
support hypothesis-driven research across all fields of life science.
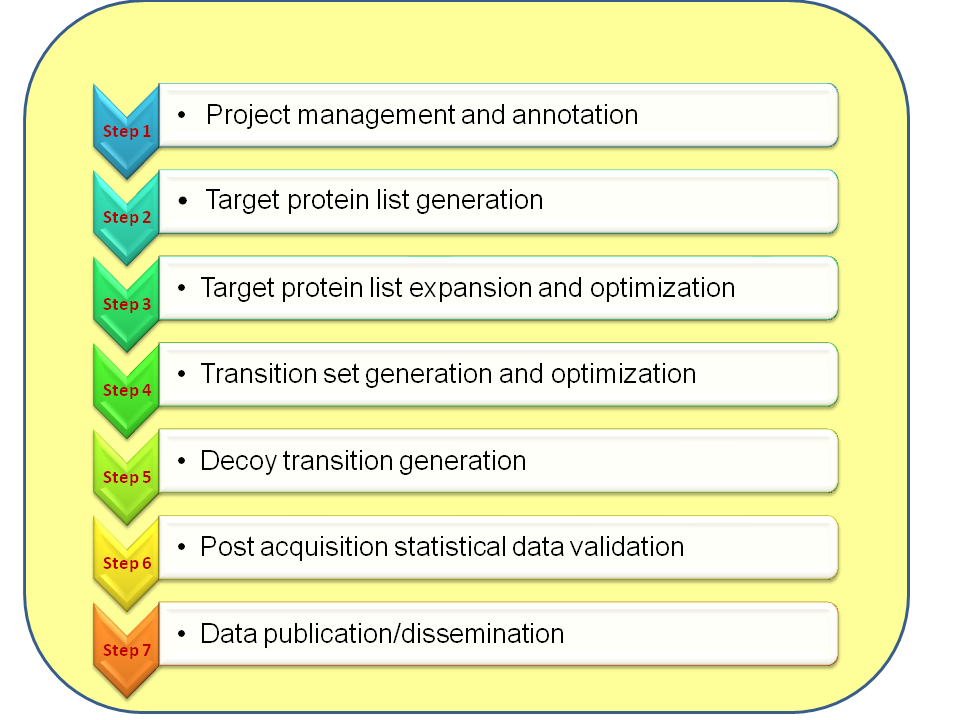
Figure. Summary of ATAQS workflow. ATAQS is composed of seven steps. The data flow is
flexible in that the user can select which steps they want to use. For example,
if user has already generated and optimized the list of transitions and decoy
transitions, then the user can skip steps 4 and 5.
ATAQS article and contributing authors
ATAQS: A Computational Software
Tool for High Throughput Transition Optimization and Validation for Selected
Reaction Monitoring Mass Spectrometry, BMC Bioinformatics 2011, 12:78 (You can download the entire article by clicking this link).
Mi-Youn K. Brusniak, Sung-Tat
Kwok, Mark Christiansen, David Campbell, Lukas Reiter, Paola Picotti, Ulrike
Kusebauch, Hector Ramos, Eric W. Deutsch, Jingchun Chen, Robert L. Moritz, Ruedi
Aebersold
ATAQS software installation instruction and
source code
You can download software installation world
document by the link ATAQS Installation Guide.doc. ATAQS_v1.0.tgz (downloadable from sourceforge.net Corra-ATAQS project) contains
complete source codes and libraries.
Corra-ATAQS developer code base and discussion group
Please join as project member for future development and biomedical research collaborators and please join for discussion.
ATAQS user guide and example data set
PDF format of ATAQS User guide using example
yeast data set in the paper can be downloaded by the link ATAQS User Guide.pdf
and ATAQS_Yeast_Example_Data_Set.zip (30 MB)
ATAQS demo site
If you would like
to take a quick look without installation in your site, you can also use our
demo website which is designed for “read only” pipeline. More specifically, it
will not allow user to launch jobs to our server. However, this site can give
you a pretty good idea on workflow and function so you can use ATAQS user guide
mentioned in the previous section.
http://ataqsdemo.systemsbiology.net
Discovery tool
While ATAQS is a computational tool for hypothesis-driven research across all fields of life science, Corra is a computational tool for discovery of candidate proteins of interest by utilizing label free quantification.
Messages:
Thank you for
interested in using ATAQS for your biological data application or enhancing
current software or algorithm research.
We are looking
forward to hearing from you and future collaboration as a community based code
development.
Sincerely,
Mi-Youn Brusniak, Ph.D.
/**
* Copyright (c) 2010 Institute for Systems Biology
*
* Licensed under the Apache License, Version 2.0 (the "License");
* you may not use this file except in compliance with the License.
* You may obtain a copy of the License at
*
* http://www.apache.org/licenses/LICENSE-2.0
*
* Unless required by applicable law or agreed to in writing, software
* distributed under the License is distributed on an "AS IS" BASIS,
* WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
* See the License for the specific language governing permissions and
* limitations under the License.
*
* Contact info:
* Mi-Youn K. Brusniak, Ph.D.
* Insitute for Systems Biology
* 1441 North 34th St.
* Seattle, WA 98103 USA
* mbrusniak@systemsbiology.org
*
*/
