TPP Tutorial v1
From SPCTools
| Revision as of 17:19, 6 July 2007 Bryanp (Talk | contribs) ← Previous diff |
Revision as of 18:20, 6 July 2007 Bryanp (Talk | contribs) Next diff → |
||
| Line 94: | Line 94: | ||
| This tutorial does not require a search engine. Searched data is provided. A computer running Windows Vista, XP or 2000 is required. Currently builds of TPP are distributed for the cygwin environment running in Windows, Linux and native Windows. This tutorial focuses on TPP run in the cygwin environment on computers running the Windows operating system. A web browser such as Firefox or Internet Explorer is required. Including the TPP software, the tutorial requires about 900MB of hard drive space. TPP itself requires approximately 190MB of disk space. The remaining space is necessary to store and manipulate the data. | This tutorial does not require a search engine. Searched data is provided. A computer running Windows Vista, XP or 2000 is required. Currently builds of TPP are distributed for the cygwin environment running in Windows, Linux and native Windows. This tutorial focuses on TPP run in the cygwin environment on computers running the Windows operating system. A web browser such as Firefox or Internet Explorer is required. Including the TPP software, the tutorial requires about 900MB of hard drive space. TPP itself requires approximately 190MB of disk space. The remaining space is necessary to store and manipulate the data. | ||
| For TPP analysis of your data it is important to remember that TPP requires that mass spectrometer data be saved in mzXML or mzDATA formats. mzXML and mzDATA are instrument independent data formats used by data analysis software like TPP and data repositories. mzXML was developed by the Institute for Systems Biology and mzData, developed by the HUPO PSI standards group. Unfortunately, storing data in both the mass spectrometer manufacture specific format and one instrument independent data format will require more than twice as much storage space for data. | For TPP analysis of your data it is important to remember that TPP requires that mass spectrometer data be saved in mzXML or mzDATA formats. mzXML and mzDATA are instrument independent data formats used by data analysis software like TPP and data repositories. mzXML was developed by the Institute for Systems Biology and mzData, developed by the HUPO PSI standards group. Unfortunately, storing data in both the mass spectrometer manufacture specific format and one instrument independent data format will require more than twice as much storage space for data. | ||
| + | |||
| + | ==About this Tutorial== | ||
| + | This guide uses the following typographical conventions: | ||
| + | '''Bold''' is used to indicate commands or steps that the user must complete. Small ''Itallics'' is use for notes that contain information that is not required to complete this tutorial. | ||
Revision as of 18:20, 6 July 2007
Trans Proteomic Pipeline (TPP) TUTORIAL
TPP V3.2.1, 2007. Note: Screenshots may vary from the TPP build you are using because the application is in development.
This document was originally assembled and edited by Bryan Prazen of Insilicos.
Contents |
Table of Contents
1 Introduction
1.1 About Trans Proteomic Pipeline
1.2 Systems Requirements
1.3 About this Tutorial
1.4 Who Should Use this Tutorial?
2 Getting Started
2.1 Downloading TPP
2.2 Installing TPP
2.3 DOS reminders
3 Tutorial Data
3.1 Getting the Tutorial Data
4 TPP Tutorial - SEQUEST data
4.1 Creating Summary HTML Files
4.2 Opening the GUI
4.2 Creating pepXML Files
4.3 PepXML Viewer
4.4 Peptide Level Analysis
4.4.1 PeptideProphet
4.4.2 XPRESS
4.4.3 ASAPRatio
4.4.4 Libra
4.4.5 Run Analysis
4.5 Evaluating the Results
4.5.1 PeptideProphet Results
4.5.2 XPRESS Results
4.5.3 ASAPRatio Results
4.5.4 Reviewing Processed Data
4.6 Protein Analysis
4.7 Exporting Data
4.8 Automation
5 X!Tandem/TPP Analysis
5.7 Analyze Peptides
6 Beyond this Tutorial
6.1 Creating mzXML Files
6.2 Setting up a TPP Account
7 Getting Help
8 Glossary
9 References
Introduction
This tutorial will cover the application of the Trans Proteomic Pipeline (TPP) for protein identification and quantitation to LC-tandem MS data. The data used in this tutorial has previously been searched with SEQUEST (Thermo Finnigan). Although this tutorial should be helpful to anyone interested in statistical identification and quantitative analysis of proteins with mass spectrometry, this tutorial was designed for the scientist who is currently running SEQUEST searches on their tandem mass spectrometry data and would like to process their data a step further. This tutorial shows an example of how to run the TPP tools so that searched data can be statistically evaluated, quantified and organized using TPP. This tutorial focuses on the application of TPP and only briefly touches on the bioinformatics behind the tools which are included in TPP.
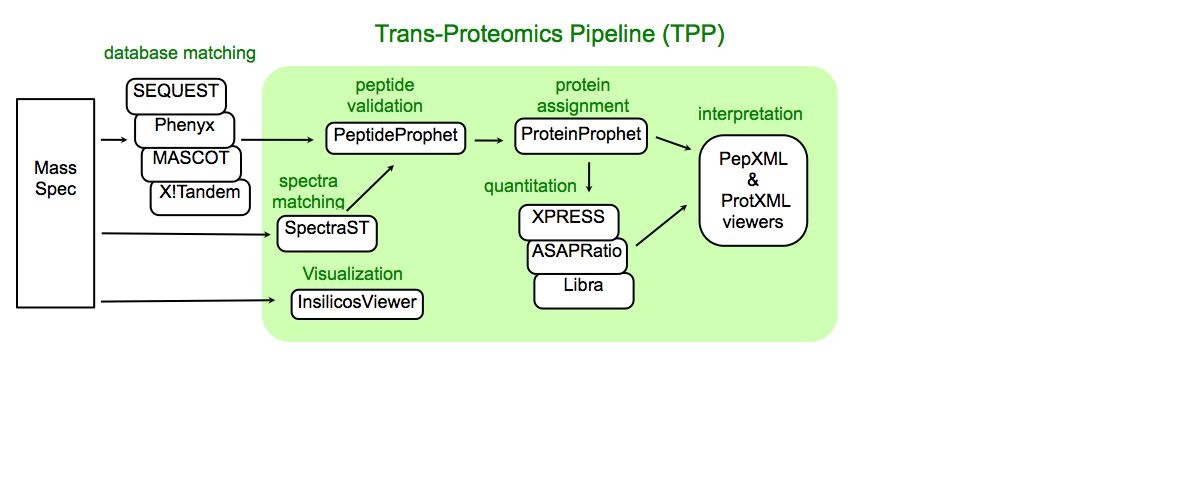
About Trans-Proteomic Pipeline
Trans-Proteomic Pipeline (TPP) is a data analysis pipeline for the analysis of LC/MS/MS proteomics data. TPP includes modules for validation of database search results, quantitation of isotopically labeled samples, and validation of protein identifications, as well as tools for viewing raw LC/MS data, peptide identification results, and protein identification results. The XML backbone of this pipeline enables a uniform analysis for LC/MS/MS data generated by a wide variety of mass spectrometer types, and assigned peptides using a wide variety of database search engines.
Systems Requirements
This tutorial does not require a search engine. Searched data is provided. A computer running Windows Vista, XP or 2000 is required. Currently builds of TPP are distributed for the cygwin environment running in Windows, Linux and native Windows. This tutorial focuses on TPP run in the cygwin environment on computers running the Windows operating system. A web browser such as Firefox or Internet Explorer is required. Including the TPP software, the tutorial requires about 900MB of hard drive space. TPP itself requires approximately 190MB of disk space. The remaining space is necessary to store and manipulate the data. For TPP analysis of your data it is important to remember that TPP requires that mass spectrometer data be saved in mzXML or mzDATA formats. mzXML and mzDATA are instrument independent data formats used by data analysis software like TPP and data repositories. mzXML was developed by the Institute for Systems Biology and mzData, developed by the HUPO PSI standards group. Unfortunately, storing data in both the mass spectrometer manufacture specific format and one instrument independent data format will require more than twice as much storage space for data.
About this Tutorial
This guide uses the following typographical conventions: Bold is used to indicate commands or steps that the user must complete. Small Itallics is use for notes that contain information that is not required to complete this tutorial.

