Software:ASAPRatio
From SPCTools
| Revision as of 18:44, 9 April 2007 Jtasman (Talk | contribs) ← Previous diff |
Current revision Jtasman (Talk | contribs) |
||
| Line 1: | Line 1: | ||
| + | [[Image:ASAPRatio_GUI.jpg|right]] | ||
| + | ==Getting the software== | ||
| '''This software is included in the current [[Software:TPP|TPP]] distribution.''' | '''This software is included in the current [[Software:TPP|TPP]] distribution.''' | ||
| + | ==In a nutshell== | ||
| Automated Statistical Analysis on Protein Ratio (ASAPRatio) | Automated Statistical Analysis on Protein Ratio (ASAPRatio) | ||
| + | ==More detail== | ||
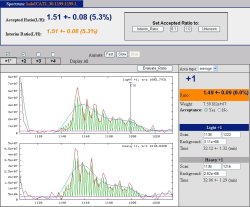
| + | '''Automated Statistical Analysis on Protein Ratio (ASAPRatio)''' accurately calculates the relative abundances of proteins and the corresponding confidence intervals from ICAT-type ESI-LC/MS data. The software first uses a Savitzky- Golay smoothing filter to reconstruct LC spectra of a peptide and its partner in a single charge state, subtracts background noise from each spectrum, and calculates light:heavy ratio of the peptide in that charge state. The ratios of the same peptide in different charge states are averaged and weighted by the corresponding spectrum intensity to obtain the peptide light:heavy ratio and its error. Subsequently, all unique peptides identified for a given protein are collected, their ratios and errors calculated, outlier's are checked for using Dixon's tests, and the relative abundance and confidence interval for the protein are calculated by applying statistics for weighed samples. The software quickly generates a list of interesting proteins based on their relative abundance, which is the goal of many proteomics experiments but was time- consuming and tedious before. A byproduct of the software is to identify outlier peptides which may be misidentified or, more interestingly, post-translationally modified. | ||
| - | Reference: Li X-J, Zhang H, Ranish JR, and Aebersold R. (2003) "Automated statistical analysis of protein abundance ratios from data generated by stable isotope dilution and tandem mass spectrometry." Anal Chem 75:6648-57. | + | ==Reference== |
| + | Li X-J, Zhang H, Ranish JR, and Aebersold R. (2003) "Automated statistical analysis of protein abundance ratios from data generated by stable isotope dilution and tandem mass spectrometry." Anal Chem 75:6648-57. | ||
| [http://tools.proteomecenter.org/publications/Li.ASAP.pdf download PDF] | [http://tools.proteomecenter.org/publications/Li.ASAP.pdf download PDF] | ||
Current revision
Contents |
Getting the software
This software is included in the current TPP distribution.
In a nutshell
Automated Statistical Analysis on Protein Ratio (ASAPRatio)
More detail
Automated Statistical Analysis on Protein Ratio (ASAPRatio) accurately calculates the relative abundances of proteins and the corresponding confidence intervals from ICAT-type ESI-LC/MS data. The software first uses a Savitzky- Golay smoothing filter to reconstruct LC spectra of a peptide and its partner in a single charge state, subtracts background noise from each spectrum, and calculates light:heavy ratio of the peptide in that charge state. The ratios of the same peptide in different charge states are averaged and weighted by the corresponding spectrum intensity to obtain the peptide light:heavy ratio and its error. Subsequently, all unique peptides identified for a given protein are collected, their ratios and errors calculated, outlier's are checked for using Dixon's tests, and the relative abundance and confidence interval for the protein are calculated by applying statistics for weighed samples. The software quickly generates a list of interesting proteins based on their relative abundance, which is the goal of many proteomics experiments but was time- consuming and tedious before. A byproduct of the software is to identify outlier peptides which may be misidentified or, more interestingly, post-translationally modified.
Reference
Li X-J, Zhang H, Ranish JR, and Aebersold R. (2003) "Automated statistical analysis of protein abundance ratios from data generated by stable isotope dilution and tandem mass spectrometry." Anal Chem 75:6648-57. download PDF