Software:Firegoose, Gaggle, and PIPE
From SPCTools
| Revision as of 17:58, 4 September 2008 Hramos (Talk | contribs) ← Previous diff |
Current revision Hramos (Talk | contribs) |
||
| Line 6: | Line 6: | ||
| ==Figures== | ==Figures== | ||
| - | [[Image:Cytoscape.png|center]] | + | [[Image:pipeWiki1.jpg|center]] |
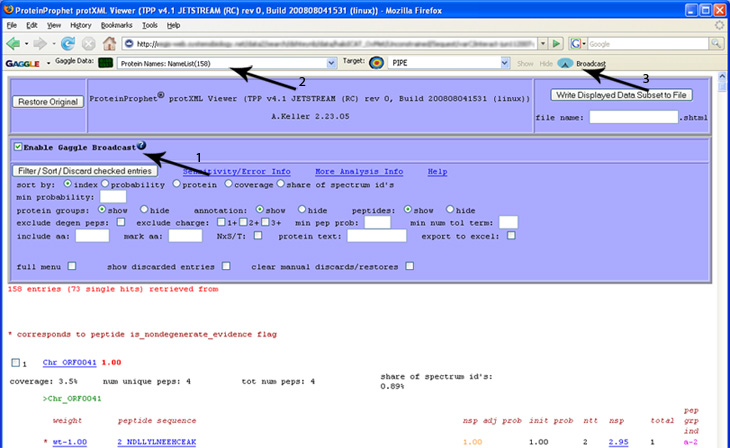
| + | 1) "Enable Gaggle Broadcast" checbox has been checked. Once this is done, 2) the Firegoose toolbar is able to read the protein list directly from the Protein Prophet page. 3) Clicking the Broadcast button sends the protein list to the selected target, in this case, the PIPE. | ||
| + | |||
| + | [[Image:pipeWiki2.jpg|center]] | ||
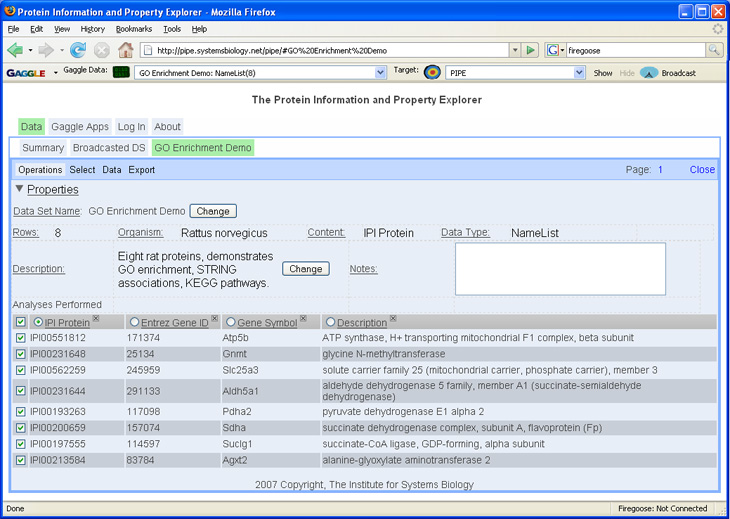
| + | Once a list has been broadcasted into the PIPE, ID mapping and lookup operations can be performed to continue analysis on the protein list. More documentation is available from the [http://pipe.systemsbiology.net/ PIPE website]. | ||
| ==Reference== | ==Reference== | ||
Current revision
Contents |
Description
The Protein Prophet output page has recently been made Gaggle compatible. This means that, if the Firegoose is installed on your Firefox browser, your lists of proteins can be directly sent to any Gaggle application, including the PIPE web application for functional annotation and analysis. The Firegoose is a Firefox pluggin that effectively turns the Firefox browser into a "Goose" that can communicate with other "Geese" as part of the Gaggle. A goose is any piece of software that has been slightly modified to accept and send small messages defined by the Gaggle standards. The Gaggle allows a user to leverage software tools developed independently and created specifically for different computational tasks in order to analyze their data through different means and gain greater insight.
The PIPE
The PIPE (Protein Information and Property Explorer) was developed as a first next-step after TPP processing of mass-spec data has been performed. Once protein identifiers have been established, the PIPE can perform several different operations on the list of IDs, including: mapping to other IDs (entrez gene, unigene, uniprot, etc.), Gene Ontology enrichment calculations (to see what biological processes are overrepresented in the list), annotation of the list to specific GO terms, and more. The PIPE can also serve as a data management tool, saving lists with user specified metadata (i.e., description of the data).
Figures
1) "Enable Gaggle Broadcast" checbox has been checked. Once this is done, 2) the Firegoose toolbar is able to read the protein list directly from the Protein Prophet page. 3) Clicking the Broadcast button sends the protein list to the selected target, in this case, the PIPE.
Once a list has been broadcasted into the PIPE, ID mapping and lookup operations can be performed to continue analysis on the protein list. More documentation is available from the PIPE website.
Reference
The Gaggle: An open-source software system for integrating bioinformatics software and data sources. Paul T Shannon, David J Reiss, Richard Bonneau, Nitin S Baliga. BMC Bioinformatics 2006, 7:176
The Protein Information and Property Explorer: an easy-to-use, rich-client web application for the management and functional analysis of proteomic data. Hector Ramos, Paul Shannon, Ruedi Aebersold. Bioinformatics, doi:10.1093/bioinformatics/btn363
The Firegoose: two-way integration of diverse data from different bioinformatics web resources with desktop applications. J Christopher Bare, Paul T Shannon, Amy K Schmid, and Nitin S Baliga. BMC Bioinformatics 2007, 8:456