Software:Prequips:Inclusion List Builder
From SPCTools
| Revision as of 23:32, 23 September 2007 Gehlenbo (Talk | contribs) ← Previous diff |
Current revision Gehlenbo (Talk | contribs) (→Software) |
||
| Line 1: | Line 1: | ||
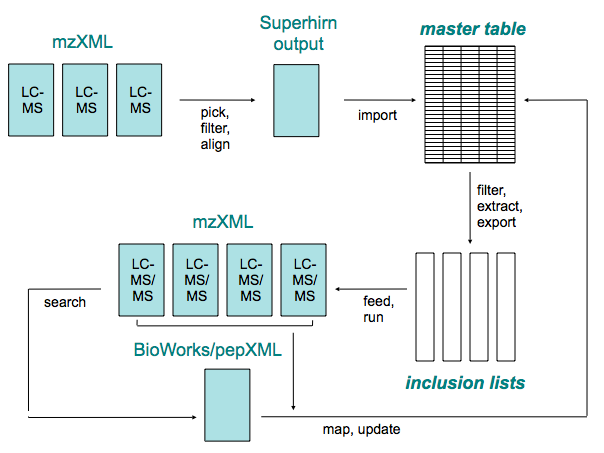
| - | = Prequips Inclusion List Builder Extension = | + | The Prequips Inclusion List Builder extension is a plug-in for [[Prequips]]. The Inclusion List Builder facilitates a directed LC-MS/MS that decouples peak detection and sequencing of selected precursor ions. The method has been described in detail in ''Schmidt A, Gehlenborg N, Bodenmiller B, Mueller L, Domon B and Aebersold R, "An integrated, directed mass spectrometric approach for in depth characterization of complex peptide mixtures." (submitted)''. |
| - | == About == | + | The Inclusion List Builders supports creation of inclusion lists and subsequent mapping of identified peptides to features. |
| - | The Prequips Inclusion List Builder Extension is a plug-in for Prequips. The Inclusion List Builder facilitates a directed LC-MS/MS that decouples peak detection and sequencing of selected precursor ions. | + | Please contact [mailto:ngehlenborg@systemsbiology.org Nils Gehlenborg] if you experience problems with the Inclusion List Builder or Prequips. |
| - | === Publications === | + | == Concept == |
| - | * Schmidt A, Gehlenborg N, Bodenmiller B, Mueller L, Domon B and Aebersold R, "An integrated, directed mass spectrometric approach for in depth characterization of complex peptide mixtures." (submitted) | + | === Overview === |
| + | [[Image:Ilb_concept.png|alt Inclusion List Builder concept]] | ||
| + | |||
| + | |||
| + | === Step-By-Step in Prequips === | ||
| + | # Create a project. | ||
| + | # Create a multi-sample analysis as part of this project. | ||
| + | # Create two single-sample analyses, say "Mapping Runs" and "INL Runs", as part of the multi-sample analysis. | ||
| + | |||
| + | To be continued. | ||
| == Download == | == Download == | ||
| === Software === | === Software === | ||
| - | We currently provide the Inclusion List Builder only bundled with Prequips. | + | We currently provide the Inclusion List Builder only bundled with [[Prequips]]. |
| - | * Download for Windows: [http://www.prequips.org/download/software/prequips_0.96_win32.zip Prequips 0.96 + Inclusion List Builder] | + | * '''NEW''' [http://www.prequips.org/download/software/prequips_0.97_win32.zip Prequips 0.97 + Inclusion List Builder] for Windows |
| - | * Download for Mac OS X: [http://www.prequips.org/download/software/prequips_0.96_macosx.zip Prequips 0.96 + Inclusion List Builder] | + | * '''NEW''' [http://www.prequips.org/download/software/prequips_0.97_macosx.zip Prequips 0.97 + Inclusion List Builder] for Mac OS X |
| === Sample Data === | === Sample Data === | ||
| + | Download the ZIP archive below for a sample data set to explore the features of the Inclusion List Builder: | ||
| + | |||
| + | [http://www.prequips.org/download/data/prequips_ilb_sample-data.zip Sample Data] (80 MB). Contains 1 mzXML file in centroid mode for the mapping runs, 1 mzXML file in centroid mode for the inclusion list runs, the corresponding peptide (and protein) identification in BioWorks tabular file format and the Superhirn (MASTER_RUN) file containing the list of detected features in the mapping runs. (Originally the mapping runs comprised 3 files and the inclusion list runs 5 files.) | ||
| == Prerequisites == | == Prerequisites == | ||
| # Make sure you have installed and configured a Java Runtime Environment (JRE) version 1.5 or later. Most machines should have this installed by default. Should your machine not have a recent JRE installed you may download a more up-to-date version from http://java.sun.com. | # Make sure you have installed and configured a Java Runtime Environment (JRE) version 1.5 or later. Most machines should have this installed by default. Should your machine not have a recent JRE installed you may download a more up-to-date version from http://java.sun.com. | ||
| - | # Prequips requires large amounts of memory. We recommend at least 1024 MB. | + | # [[Prequips]] requires large amounts of memory. We recommend at least 1024 MB. |
| == Installation == | == Installation == | ||
| Line 30: | Line 42: | ||
| # Execute the Prequips application (prequips.app, prequips.exe, ...). A splash screen should appear within a few seconds. | # Execute the Prequips application (prequips.app, prequips.exe, ...). A splash screen should appear within a few seconds. | ||
| - | == Screenshot Gallery == | + | == Tutorial Screencasts == |
| - | The screenshots in this gallery are meant to give potential users an impression of what the user interface will look like (even though it's still in beta). This is by no means a complete overview of all features and the presented features are in shown in no particular order. | + | These tutorials require the free Apple Quicktime Player: http://www.apple.com/quicktime. All five parts together are a basic walk-through the functionality of the Inclusion List Builder. |
| + | |||
| + | # [http://www.prequips.org/download/docs/prequips_setup-project.mov How to create a new project.] | ||
| + | # [http://www.prequips.org/download/docs/prequips_create-master-table.mov How to create the master table.] | ||
| + | # [http://www.prequips.org/download/docs/prequips_create-inclusion-list.mov How to create an inclusion list.] | ||
| + | # [http://www.prequips.org/download/docs/prequips_map-peptides-to-master-table.mov How map identified peptides back to the master table.] | ||
| + | # [http://www.prequips.org/download/docs/prequips_export-peptide-mappings.mov How to export the peptide mappings of the master table.] | ||
| + | |||
| + | == Screenshot Gallery == | ||
| <gallery> | <gallery> | ||
| - | image:Load_alignment.png|Loading an alignment using a so-called "alignment provider". Currently there are alignment providers for Superhirn (tabular format)and SpecArray. Providers for other tools or formats (i.e. Superhirn XML format) can be added easily (once they are implemented ...). | + | image:Ilb_basics_01.png|Master table, filters and feature details. |
| - | image:Create_inclusion_list.png|Create a new inclusion list from the features shown in the master table (big table on the top right). Filters (as shown on the bottom right) are used to select only features fulfilling specified criteria from the overall list of features. | + | image:Ilb_basics_03.png|Segment information, inclusion list overview and exported inclusion list. |
| - | image:Create_segments.png|Assign feature in selected inclusion list to segments. | + | image:Ilb_basics_02.png|Master table and feature details with mapping information. |
| - | image:Edit_charge_filter.png|Modify the settings of a filter. A filter can also be negated. | + | |
| - | image:Edit_meta_filter.png|Modify the mode of a "meta filter". A meta filter combines several regular filters using a logical operator (AND, OR, XOR). There can be additional meta filters (using different operators). However, meta filters are always combined using AND, i.e. a feature has to pass every meta filter to be included in the filtered list. | + | |
| - | image:Inclusion_list_overview.png|All inclusion lists created from the master table, which is represented by the top entry called "Superhirn Alignment" (technically the master table is an inclusion list as well). Some preliminary statistics are shown. This will be extended. | + | |
| </gallery> | </gallery> | ||
Current revision
The Prequips Inclusion List Builder extension is a plug-in for Prequips. The Inclusion List Builder facilitates a directed LC-MS/MS that decouples peak detection and sequencing of selected precursor ions. The method has been described in detail in Schmidt A, Gehlenborg N, Bodenmiller B, Mueller L, Domon B and Aebersold R, "An integrated, directed mass spectrometric approach for in depth characterization of complex peptide mixtures." (submitted).
The Inclusion List Builders supports creation of inclusion lists and subsequent mapping of identified peptides to features.
Please contact Nils Gehlenborg if you experience problems with the Inclusion List Builder or Prequips.
Contents |
Concept
Overview
Step-By-Step in Prequips
- Create a project.
- Create a multi-sample analysis as part of this project.
- Create two single-sample analyses, say "Mapping Runs" and "INL Runs", as part of the multi-sample analysis.
To be continued.
Download
Software
We currently provide the Inclusion List Builder only bundled with Prequips.
- NEW Prequips 0.97 + Inclusion List Builder for Windows
- NEW Prequips 0.97 + Inclusion List Builder for Mac OS X
Sample Data
Download the ZIP archive below for a sample data set to explore the features of the Inclusion List Builder:
Sample Data (80 MB). Contains 1 mzXML file in centroid mode for the mapping runs, 1 mzXML file in centroid mode for the inclusion list runs, the corresponding peptide (and protein) identification in BioWorks tabular file format and the Superhirn (MASTER_RUN) file containing the list of detected features in the mapping runs. (Originally the mapping runs comprised 3 files and the inclusion list runs 5 files.)
Prerequisites
- Make sure you have installed and configured a Java Runtime Environment (JRE) version 1.5 or later. Most machines should have this installed by default. Should your machine not have a recent JRE installed you may download a more up-to-date version from http://java.sun.com.
- Prequips requires large amounts of memory. We recommend at least 1024 MB.
Installation
- Extract the downloaded Prequips ZIP archive into a directory of your choice. A new subdirectory named "prequips_xxx" will be created ("xxx" will be replaced by the version and the OS identifier, e.g. "macosx" or "win32").
- Execute the Prequips application (prequips.app, prequips.exe, ...). A splash screen should appear within a few seconds.
Tutorial Screencasts
These tutorials require the free Apple Quicktime Player: http://www.apple.com/quicktime. All five parts together are a basic walk-through the functionality of the Inclusion List Builder.
- How to create a new project.
- How to create the master table.
- How to create an inclusion list.
- How map identified peptides back to the master table.
- How to export the peptide mappings of the master table.