TPP Tutorial v1
From SPCTools
| Revision as of 23:55, 6 July 2007 Bryanp (Talk | contribs) (→Creating pepXML Files) ← Previous diff |
Revision as of 23:56, 6 July 2007 Bryanp (Talk | contribs) (→Creating pepXML Files) Next diff → |
||
| Line 237: | Line 237: | ||
| Such as: | Such as: | ||
| + | |||
| Sequest2XML raft4041.html -Psequest.params | Sequest2XML raft4041.html -Psequest.params | ||
| + | |||
| Sequest2XML raft4243.html -Psequest.params | Sequest2XML raft4243.html -Psequest.params | ||
| + | |||
| etc. | etc. | ||
| ''NOTE: When analyzing your own data, the working directory must contain the .html and .mzXML as well as the SEQUEST results in .tgz or subdirectorys for Sequest2XML to work.'' | ''NOTE: When analyzing your own data, the working directory must contain the .html and .mzXML as well as the SEQUEST results in .tgz or subdirectorys for Sequest2XML to work.'' | ||
Revision as of 23:56, 6 July 2007
Trans Proteomic Pipeline (TPP) Tutorial
TPP V3.2.1, 2007. Note: Screenshots may vary from the TPP build you are using because the application is in development.
This document was originally assembled and edited by Bryan Prazen of Insilicos.
Contents |
Introduction
This tutorial will cover the application of the Trans Proteomic Pipeline (TPP) for protein identification and quantitation to LC-tandem MS data. The data used in this tutorial has previously been searched with SEQUEST (Thermo Finnigan). Although this tutorial should be helpful to anyone interested in statistical identification and quantitative analysis of proteins with mass spectrometry, this tutorial was designed for the scientist who is currently running SEQUEST searches on their tandem mass spectrometry data and would like to process their data a step further. This tutorial shows an example of how to run the TPP tools so that searched data can be statistically evaluated, quantified and organized using TPP. This tutorial focuses on the application of TPP and only briefly touches on the bioinformatics behind the tools which are included in TPP.
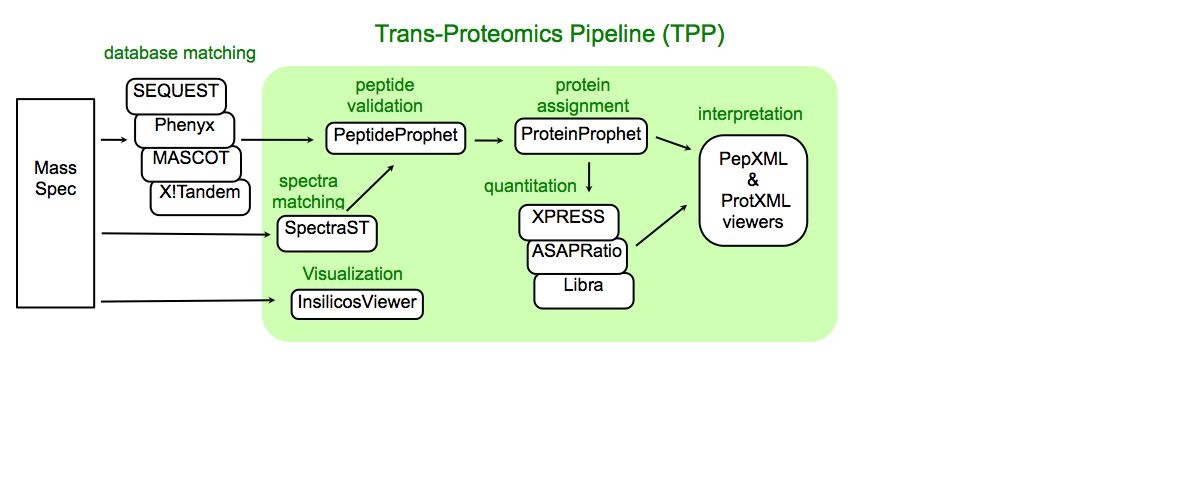
About Trans-Proteomic Pipeline
Trans-Proteomic Pipeline (TPP) is a data analysis pipeline for the analysis of LC/MS/MS proteomics data. TPP includes modules for validation of database search results, quantitation of isotopically labeled samples, and validation of protein identifications, as well as tools for viewing raw LC/MS data, peptide identification results, and protein identification results. The XML backbone of this pipeline enables a uniform analysis for LC/MS/MS data generated by a wide variety of mass spectrometer types, and assigned peptides using a wide variety of database search engines.
Systems Requirements
This tutorial does not require a search engine. Searched data is provided. A computer running Windows, XP or 2000 is required. Currently builds of TPP are distributed for the cygwin environment running in Windows, Linux and native Windows. This tutorial focuses on TPP run in the cygwin environment on computers running the Windows operating system. A web browser such as Firefox or Internet Explorer is required. Including the TPP software, the tutorial requires about 900MB of hard drive space. TPP itself requires approximately 190MB of disk space. The remaining space is necessary to store and manipulate the data. For TPP analysis of your data it is important to remember that TPP requires that mass spectrometer data be saved in mzXML or mzDATA formats. mzXML and mzDATA are instrument independent data formats used by data analysis software like TPP and data repositories. mzXML was developed by the Institute for Systems Biology and mzData, developed by the HUPO PSI standards group. Unfortunately, storing data in both the mass spectrometer manufacture specific format and one instrument independent data format will require more than twice as much storage space for data.
About this Tutorial
This guide uses the following typographical conventions: Bold is used to indicate commands or steps that the user must complete. Small Itallics is use for notes that contain information that is not required to complete this tutorial.
Who Should Use this Tutorial?
This tutorial is written for anyone who has a general interest in learning about one method to identify and quantify peptides and proteins using mass spectrometry. We have attempted to write this tutorial so that the user does not need an extraordinary knowledge of proteomics, biology, chemistry, mass spectrometry, or software engineering. Also, this tutorial does not require any software or data that is not easily available on the web and it does not require any previous experience with the analysis of mass spectrometric data. This tutorial should also be of use to those who are very familiar with proteomics data analysis but do not have a great deal of experience with TPP.
Getting Started
Downloading and Installing TPP
Information on installing and downloading the Windows Cygwin distribution of TPP can be found at: TPP:Windows_Cygwin_Installation
Getting familiar with Cygwin
The TPP GUI nearly eliminates the need to use the command line, but this section is included in the tutorial because the the Windows Cygwin distribution is running in a Cygwin environment and we do not want you to be totally lost if at some point you must step out of the GUI environment and type some commands.
Cygwin is a Linux-like environment for Windows that consists of a program which acts as a Linux interface emulation and a collection of tools, which provide Linux look and feel. Cygwin is aimed mainly at porting software that runs on Linux systems to run on Windows with only minor software changes. Cygwin is installed with the TPP during the installation procedure.
After installing the TPP, the Cygwin Bash Shell can be found under Cygwin in the Windows start menu. If you are a little familiar with Linux, Unix, or DOS you will not have any problem running TPP from Cygwin.
Below is a short list of commands for the Cygwin shell that will help you find your way around the Cygwin environment.
ls lists the files in a directory man displays the reference manual page about a command cd change directory; cd .. moves you backwards to the next higher subdirectory level chmod changes the permissions for a file
To cut text from the Cygwin shell first highlight the text with the mouse. Then press return, or put the mouse over the Cygwin shell window bar, right click, select edit, and then select copy. To paste text put the cursor in the desired location, put the mouse over the Cygwin shell window bar, right click, select edit and then select paste.
Wildcards
- and ? are wildcard commands in the Cygwin shell.
For example the command
ls raft4???.html
lists all the .html files in the directory that start with raft4 and have 3 characters after the ‘4’ and before the ‘.’.
The * wildcard is more general. It matches zero or any number of characters, except that it will not match a period that is the first character of a name.
ls raft4041.*
Lists all the files that start with ‘raft4041.’. Wildcards can be used in most Cygwin shell commands.
man followed by a command is a command to get the manual entry describing the command. For example man ls leads to a manual that explains the command ls in more detail than you would ever want to know. Use the spacebar to page through the manual and the q key to quit the manual.
Directory path delimiter
Directory path delimiters can be a confusing part of using Cygwin. Cygwin writes paths with / and dos or windows writes paths with \. For instance a directory in dos or windows that is C:\Inetpub\wwwroot\tutorial will be cygdrive/c/Inetpub/wwwroot/tutorial in the Cygwin shell. Many directory related commands like moving or deleting files can be done in the Windows or Cygwin environment. In this tutorial the Cygwin command will be written out to make you more familiar with Cygwin shell.
If you run into trouble operating Cygwin a Cygwin user guide can be found at Cygwin.
NOTE: The cygwin environment has many tools available which might enhance your Cygwin experience which are not included in the TPP install. These can be downloaded at the Cygwin web site (www.cygwin.com) or obtained by selecting a site other than tools.proteomecenter.org when operating the Cygwin setup program. Installation of new tools should not cause a problem with the TPP, but you should be cautious when updating versions of tools that came with the TPP installation. For instance, Gnuplot comes in a few flavors and the one we install is not the package distributed with Cygwin and has a slightly different syntax. Installing the Cygwin Gnuplot package will likely break the graph images generated by the TPP.
Setting up an account
The TPP GUI comes with one user account. This account has ‘guest’ as both the user name and password. Below are instructions for making another account.
Open the Cygwin shell and type:
cd /cygdrive/c/Inetpub/tpp-bin/users/
mkdir tutorial
cd tutorial
and
crypt isbTPPspc TPP > .password
chmod -R 777 /cygdrive/c/Inetpub/tpp-bin/users/tutorial
You have just created the password ‘TPP’ for the user ‘tutorial.’
In order to add a different username, create a tpp-bin/users/NEWUSER/ directory and run crypt isbTPPspc NEWPASSWORD > .password from this directory. In these examples "isbTPPspc" is the crypt key. This can be changed.
Tutorial Data
Getting the Tutorial Data
This tutorial uses a data set containing proteins that co-purified with lipid raft plasma membrane domains isolated from control and stimulated Jurkat human T cells. The analysis of similar data can be found in:
“The Application of New Software Tools to Quantitative Protein Profiling Via Isotope-coded Affinity Tag (ICAT) and Tandem Mass Spectrometry: II. Evaluation of Tandem Mass Spectrometry Methodologies for Large-Scale Protein Analysis, and the Application of Statistical Tools for Data Analysis and Interpretation” Priska D. von Haller, Eugene Yi, Samuel Donohoe, Kelly Vaughn, Andrew Keller, Alexey I. Nesvizhskii, Jimmy Eng, Xiao-jun Li, David R. Goodlett, Ruedi Aebersold, and Julian D. Watts, Mol Cell Proteomics 2003 2: 428-442."
The data used in this tutorial is not the same data that is described in the publication but the same scientists collected it using the same sample preparation and mass spectrometry procedures. Analysis was done on a LCQ Classic. The samples were ICAT labeled (Old-ICAT, light tag = d0 442, heavy tag = d8 450), separated by cation exchange chromatography, purified by avidin cartrages, separated by μLC, and measured with MS/MS. The tandem mass spectra were then analyzed using SEQUEST. This tutorial begins with the analysis of the SEQUEST results. Only a portion of the data from the raft experiment is used in this tutorial in order to save time and hard drive space. This tutorial uses data that has already been searched by SEQUEST so that the user does not need to have a SEQUEST license for the computer that is used for this tutorial.
Download this data at:
www.insilicos.com/spctools/spctools/data/tutorial.tgz
Tell your browser to save the file to C:\Inetpub\wwwroot\ISB\data\tutorial. You will need to create the tutorial directory.
Unpacking and Storing the TPP Tutorial Data
It is important that all the data that is analyzed with the TPP be stored in specific locations. The TPP can only see data that is located under the C:\Inetpub\wwwroot directory.
If you have the program Winzip on your computer, open the tutorial.tgz file with Winzip and extract the contents to the C:\Inetpub\wwwroot\ISB\data\tutorial directory. After successfully extracting the data delete the tutorial.tgz file.
If you do not have Winzip, open a Cygwin terminal from the Windows start menu and type the commands cd /cygdrive/c/inetpub/wwwroot/ISB/data/tutorial and tar xzvf tutorial.tgz. At this point we can erase the compressed file. Type: rm tutorial.tgz
For this tutorial and future data analysis all data should be stored in C:\Intetpub\wwwroot\ISB\data\. Each experiment can be stored in an individual folder at this location, such as our tutorial folder.
You should now have a folder named ‘tutorial’ which contains mzXML data for 6 LC runs, folders that contain the .out and .dta files, a sequest.params file and a folder containing a FASTA database.
NOTE: To analyze data from your own experiments you will need to search the data, compress the search results and convert the raw data to mzXML format. These steps are covered in the last section - Beyond this Tutorial. Moving the dbase folder to C:\cygwin\:
I suspect that you can do this in the Windows environment easily enough, but to get familiar with Cygwin let’s do it there. Open a Cygwin shell from the Windows start menu and type the following commands:
cd /cygdrive/c/inetpub/wwwroot/ISB/data/tutorial and
mv dbase /cygdrive/c/cygwin/
Many problems with TPP are associated with file permissions and these problems seem to be very machine dependent. We will start by changing the permissions of your data folder. Type the following command in the Cygwin shell:
chmod -R 777 /cygdrive/c/Inetpub/wwwroot/ISB/data/tutorial
chmod -R 777 /cygdrive/c/cygwin/dbase
For other permission related problems type the same command with the appropriate directory inserted.
SEQUEST data analysis
Creating Summary HTML Files
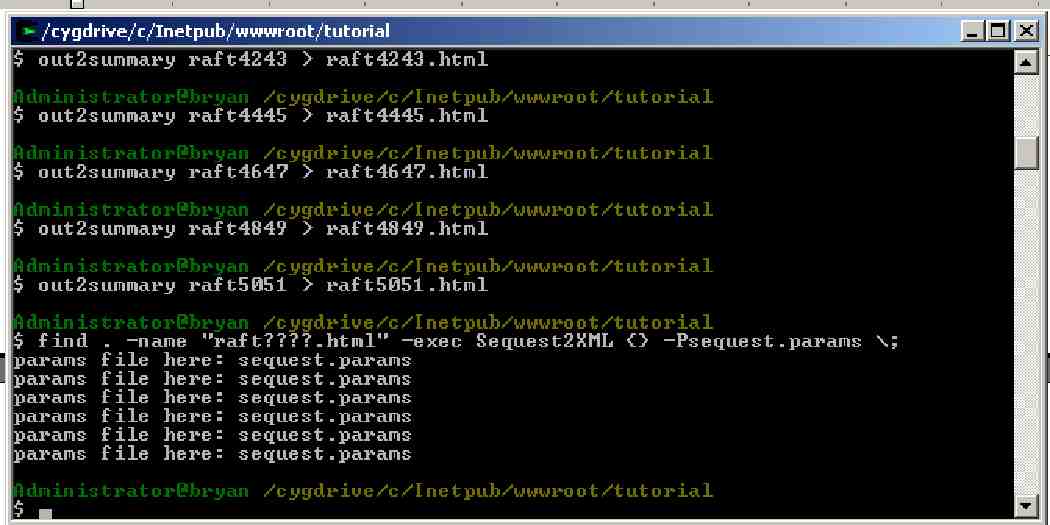
Before we get started with the GUI we will need to run one function outside the GUI. This is because the GUI assumes that a SEQUEST search will be done from the GUI. Because we decided not to require SEQUEST for this tutorial, we will first transfer the tutorial’s search results to a format the GUI can read using a text command. Each tandem mass spectrum resulting from a liquid chromatography (LC) experiment results in an individual .out file after analysis with SEQUEST or TurboSEQUEST. The first step in analyzing the tutorial results is to collect the result from a given LC separation. The Out2Summary program collates the .out files into a single HTML file for each LC separation. The original raft data contains 24 separate LC separations. For speed and portability reasons this tutorial will only analyze 6 of the 24 LC separations. The data from these 6 separations will be combined and analyzed as one single experiment.
The first step in the analysis is to change the directory in the Cygwin shell to your working directory for the tutorial. Type or copy the following command into the Cygwin shell.
cd /cygdrive/c/Inetpub/wwwroot/ISB/data/tutorial
NOTE: If you have not strayed from these instructions you will already be in the directory.
Out2Summary must be run for each LC separation. Type or copy (yes, you can copy multiple commands at once):
out2summary raft4041 > raft4041.html
out2summary raft4243 > raft4243.html
out2summary raft4445 > raft4445.html
out2summary raft4647 > raft4647.html
out2summary raft4849 > raft4849.html
out2summary raft5051 > raft5051.html
This process will take a few minutes.
NOTE: The “>” command directs output that would otherwise go to the screen to the file named raftXXXX.html
NOTE: In future analyses the base name used for the .html should match the base name used for the mzXML data (as above), if you want the instrument information to be passed to the TPP tools.
Opening the GUI
The TPP pipeline GUI can be opened by clicking on the ‘TPP Web Tools’ shortcut that was created on your desktop during installation or by selecting “TPP Web Tools” under “TPP” in the Windows start menu. Alternatively, you can click on the following link or open your favorite web browser and paste this link into the navigation bar:
http://localhost:1441/tpp-bin/tpp_gui.pl
Login as ‘tutorial’ and use ‘TPP’ as the password.
This tutorial is written from the point of view of a researcher viewing data on the computer where the TPP tools are running.
At this point you will be in the “Home” tab of the proteomics pipeline GUI. The Home tab contains information about TPP and the structure of the GUI, along with a pull down menu that lets you choose between SEQUEST, Mascot or SpectaST. The default is SEQUEST which is what will start with in this tutorial. Thus, no input is necessary under this tab.
Creating pepXML Files
For this tutorial we begin with data that has already been searched with SEQUEST so that the tutorial is instrument independent and does not require software beyond TPP. The SEQUEST Search results are in the form of .out files. In an earlier step we converted the search data in to .html files. The .html files are are vestiges of an earlier data analysis pipeline and currently only serve as an intermediate file format on the way to pepXML. TPP will analysis the search results in pepXML format. The next step will be to convert the .html files to pepXML files.
Click on “Analysis Pipeline”. This will display six tabs which activate different parts of the pipeline. The first tab is Home, which contains information about the IPP. The second tab is used to convert data from different spectrometers into mzXML, and the third tab is used to search the data. We will start with the fourth tab. Your next step is to convert the search results from .html to the pepXML format. pepXML is a file format for storing the results of database search at the peptide level. A great thing about pepXML is that its format is independent of the instrument manufacuture and database matching software. pepXML converters are currently available for SEQUEST, Mascot, COMET and X!Tandem results. Also, the Mascot software contains a pepXML exporter.
NOTE: In the near future look for the mzIdent file format that will be a Human Proteome Organisation (HUPO) standard based on pepXML.
Select the ‘pepXML’ tab in the GUI interface.
Select the ‘Add Files’ button.
Using the directory selector on the right side, navigate to the tutorial directory.
Select ‘View’ for one of the .html files.
This command opens another window that contains the SEQUEST search results for all of the spectra in a given LC run.
NOTE: This window can also be accessed at http://localhost:1441/ISB/data/Tutorial/raft4041.html.
Go back to the Main GUI page.
Check the select box to the left of each of the 6 .html files
Press the ‘Select’ button.
In the updated window,
Press ‘Add Files’ under the ‘Specify Sequest Parameters File’ section.
Check the sequest.params file and press ‘Select’.
There is no need to select any of the options and the enzyme should already be set as trypsin.
Press ‘Convert to PepXML’.
This command will take a moment to run. You will need to update the page by clicking the text “UPDATE THIS PAGE”. When the command is completed you will get 6 copies of the message “Command Successful”
At this point you have successfully converted you r search results to the pepXML format and you are ready to evaluate your data with the tools that are included in IPP. You should be aware that the same command that you just ran through the GUI could have been run from the command line. From the DOS shell and in directory raft directory you can type: Sequest2XML <file_name.html> -Psequest.params for each of the six raft????.html files in the raft directory.
Such as:
Sequest2XML raft4041.html -Psequest.params
Sequest2XML raft4243.html -Psequest.params
etc.
NOTE: When analyzing your own data, the working directory must contain the .html and .mzXML as well as the SEQUEST results in .tgz or subdirectorys for Sequest2XML to work.