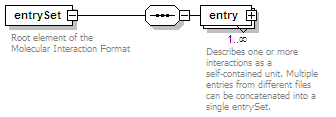
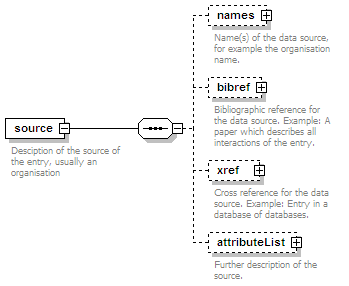
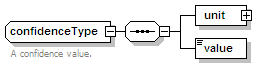| source |
<xs:complexType name="interactionElementType">
<xs:annotation>
<xs:documentation>A molecular interaction.</xs:documentation>
</xs:annotation>
<xs:sequence>
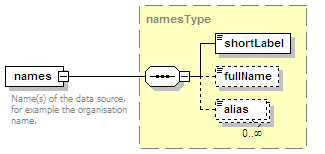
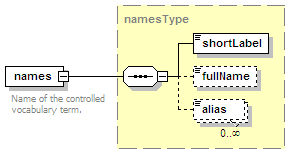
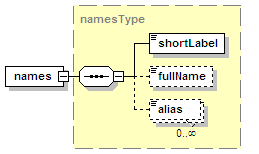
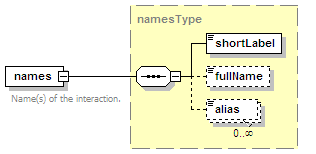
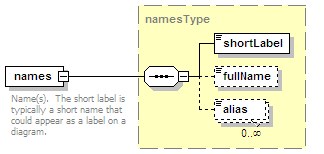
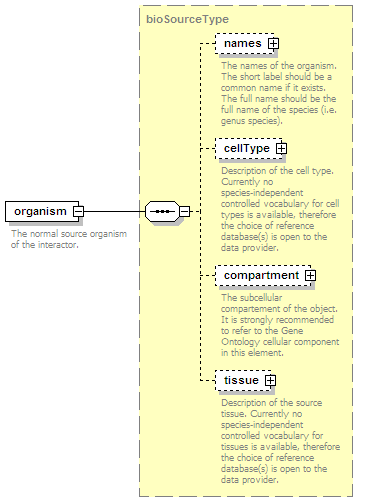
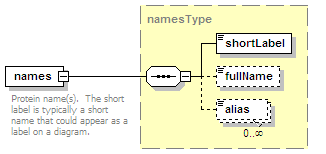
<xs:element name="names" type="namesType" minOccurs="0">
<xs:annotation>
<xs:documentation>Name(s) of the interaction.</xs:documentation>
</xs:annotation>
</xs:element>
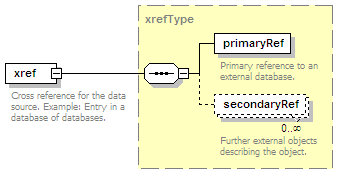
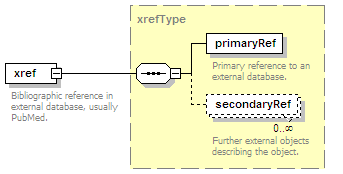
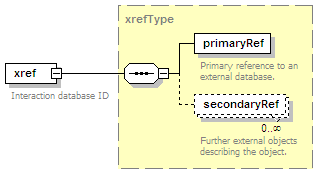
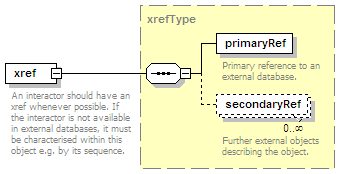
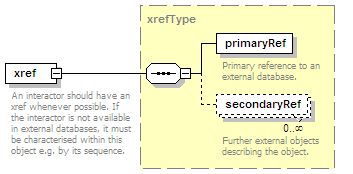
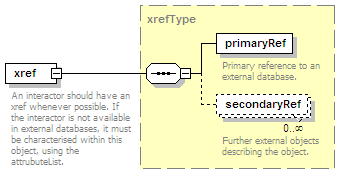
<xs:element name="xref" type="xrefType" minOccurs="0">
<xs:annotation>
<xs:documentation>Interaction database ID</xs:documentation>
</xs:annotation>
</xs:element>
<xs:choice minOccurs="0">
<xs:annotation>
<xs:documentation>Either refer to an already defined availability statement in this entry or insert description.</xs:documentation>
</xs:annotation>
<xs:element name="availabilityRef" type="refType">
<xs:annotation>
<xs:documentation>References an availability statement already present in this entry.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="availability" type="availabilityType">
<xs:annotation>
<xs:documentation>Describes the availability of the interaction data. If no availability is given, the data is assumed to be freely available.</xs:documentation>
</xs:annotation>
</xs:element>
</xs:choice>
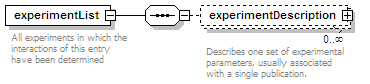
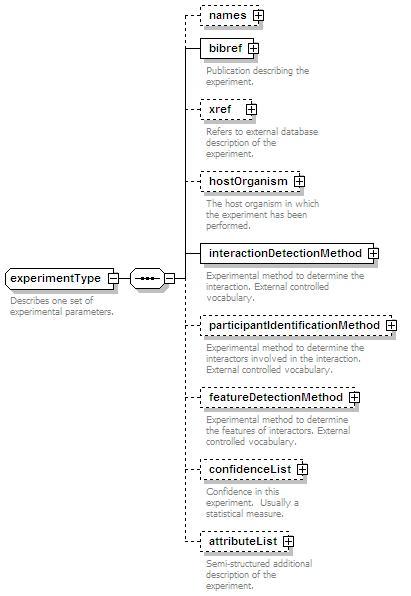
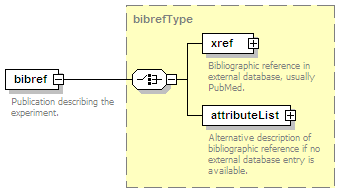
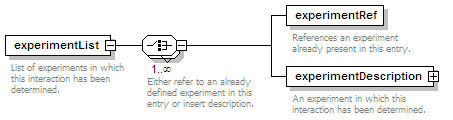
<xs:element name="experimentList">
<xs:annotation>
<xs:documentation>List of experiments in which this interaction has been determined.</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:choice maxOccurs="unbounded">
<xs:annotation>
<xs:documentation>Either refer to an already defined experiment in this entry or insert description.</xs:documentation>
</xs:annotation>
<xs:element name="experimentRef" type="refType">
<xs:annotation>
<xs:documentation>References an experiment already present in this entry.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="experimentDescription" type="experimentType">
<xs:annotation>
<xs:documentation>An experiment in which this interaction has been determined.</xs:documentation>
</xs:annotation>
</xs:element>
</xs:choice>
</xs:complexType>
</xs:element>
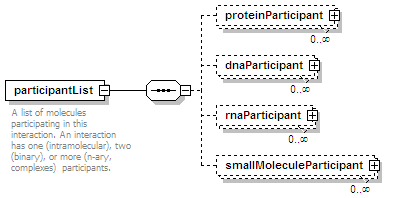
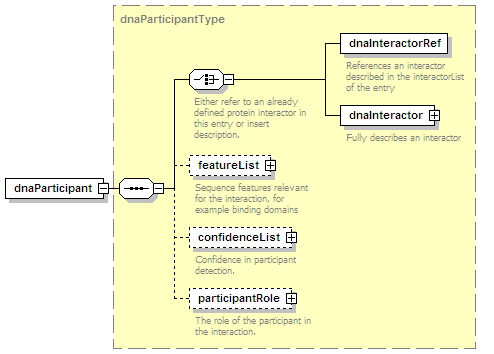
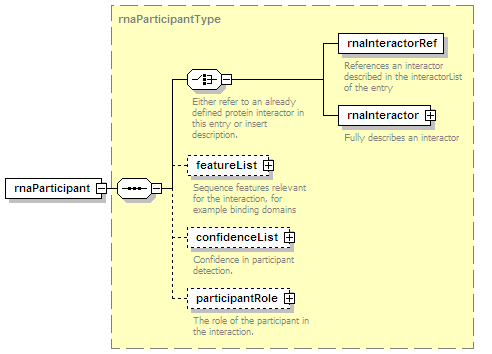
<xs:element name="participantList">
<xs:annotation>
<xs:documentation>A list of molecules participating in this interaction. An interaction has one (intramolecular), two (binary), or more (n-ary, complexes) participants.</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:sequence>
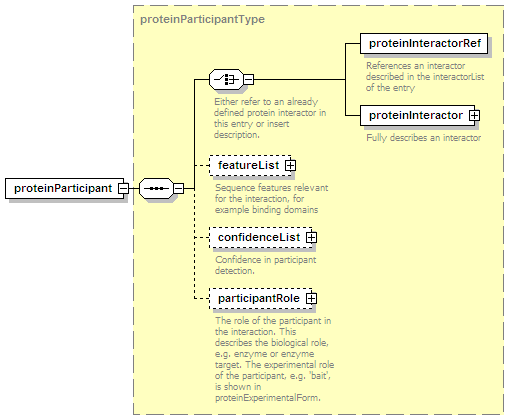
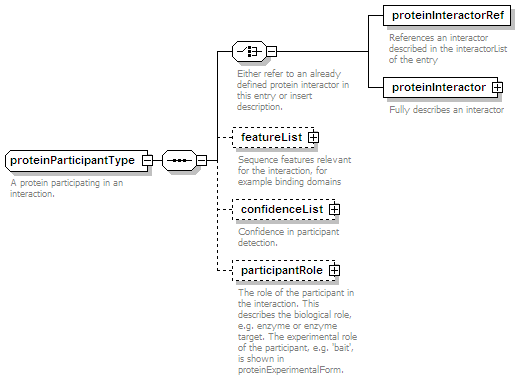
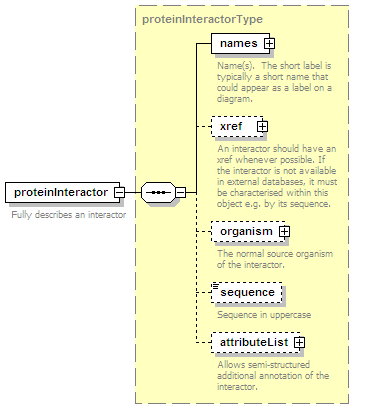
<xs:element name="proteinParticipant" minOccurs="0" maxOccurs="unbounded">
<xs:complexType>
<xs:complexContent>
<xs:extension base="proteinParticipantType"/>
</xs:complexContent>
</xs:complexType>
</xs:element>
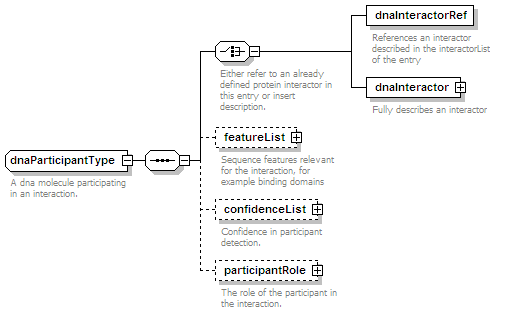
<xs:element name="dnaParticipant" type="dnaParticipantType" minOccurs="0" maxOccurs="unbounded"/>
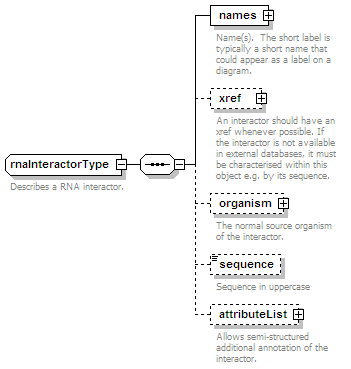
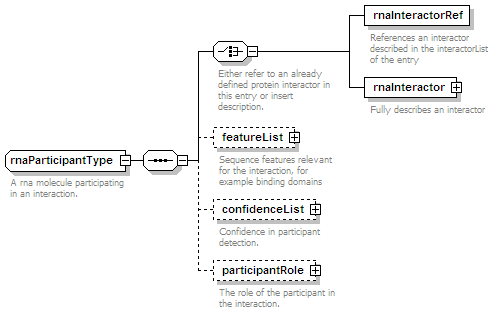
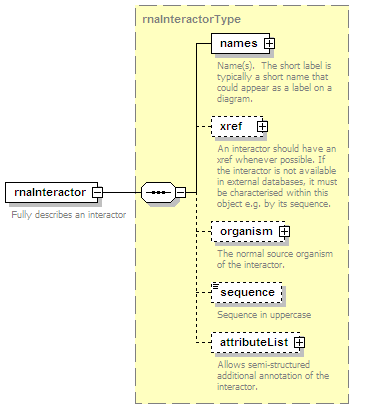
<xs:element name="rnaParticipant" type="rnaParticipantType" minOccurs="0" maxOccurs="unbounded"/>
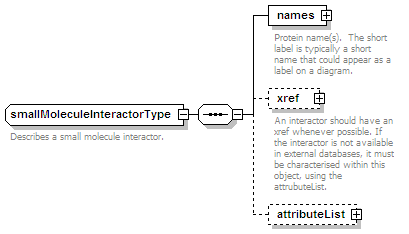
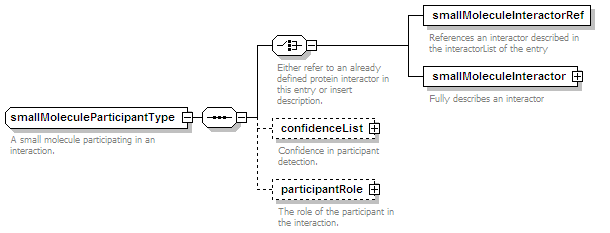
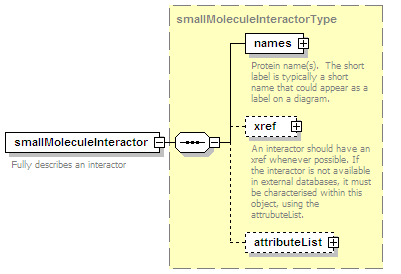
<xs:element name="smallMoleculeParticipant" type="smallMoleculeParticipantType" minOccurs="0" maxOccurs="unbounded"/>
</xs:sequence>
</xs:complexType>
</xs:element>
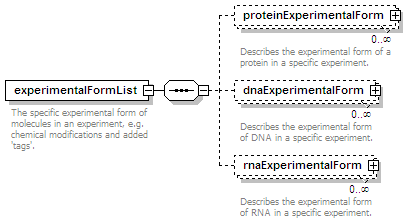
<xs:element name="experimentalFormList" minOccurs="0">
<xs:annotation>
<xs:documentation>The specific experimental form of molecules in an experiment, e.g. chemical modifications and added 'tags'.</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:sequence>
<xs:element name="proteinExperimentalForm" minOccurs="0" maxOccurs="unbounded">
<xs:annotation>
<xs:documentation>Describes the experimental form of a protein in a specific experiment. </xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:sequence>
<xs:element name="experimentRef" type="refType">
<xs:annotation>
<xs:documentation>This experimental form has been used in the experiment referred to.</xs:documentation>
</xs:annotation>
</xs:element>
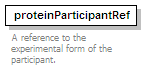
<xs:element name="proteinParticipantRef" type="refType">
<xs:annotation>
<xs:documentation>A reference to the experimental form of the participant.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="isTagged" type="xs:boolean" minOccurs="0">
<xs:annotation>
<xs:documentation>True if the protein has been tagged in the experiment.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="isOverexpressed" type="xs:boolean" minOccurs="0">
<xs:annotation>
<xs:documentation>True if the protein has been overexpressed in the experiment.</xs:documentation>
</xs:annotation>
</xs:element>
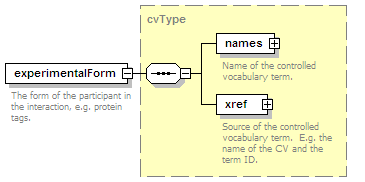
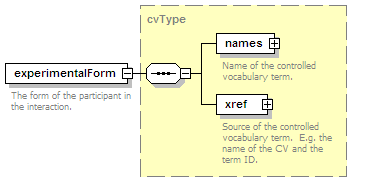
<xs:element name="experimentalForm" type="cvType" minOccurs="0">
<xs:annotation>
<xs:documentation>The form of the participant in the interaction, e.g. protein tags.</xs:documentation>
</xs:annotation>
</xs:element>
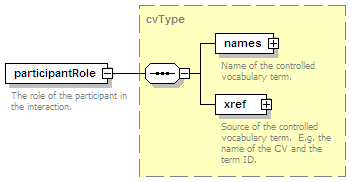
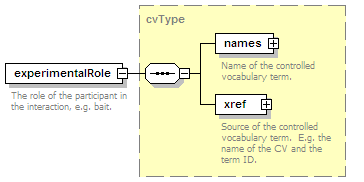
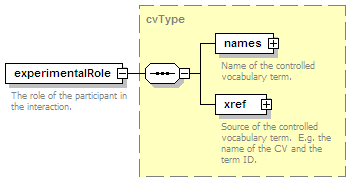
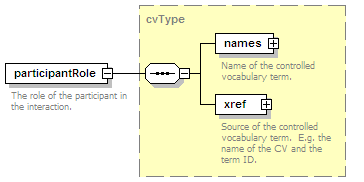
<xs:element name="experimentalRole" type="cvType" minOccurs="0">
<xs:annotation>
<xs:documentation>The role of the participant in the interaction, e.g. bait.</xs:documentation>
</xs:annotation>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
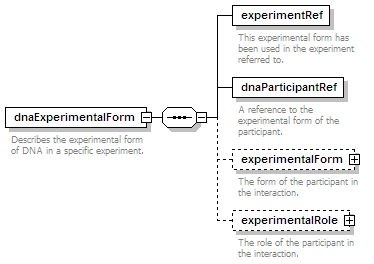
<xs:element name="dnaExperimentalForm" minOccurs="0" maxOccurs="unbounded">
<xs:annotation>
<xs:documentation>Describes the experimental form of DNA in a specific experiment.</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:sequence>
<xs:element name="experimentRef" type="refType">
<xs:annotation>
<xs:documentation>This experimental form has been used in the experiment referred to.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="dnaParticipantRef" type="refType">
<xs:annotation>
<xs:documentation>A reference to the experimental form of the participant.</xs:documentation>
</xs:annotation>
</xs:element>
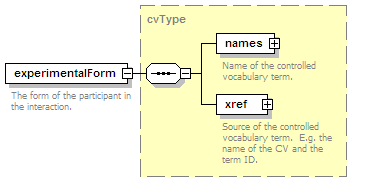
<xs:element name="experimentalForm" type="cvType" minOccurs="0">
<xs:annotation>
<xs:documentation>The form of the participant in the interaction.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="experimentalRole" type="cvType" minOccurs="0">
<xs:annotation>
<xs:documentation>The role of the participant in the interaction.</xs:documentation>
</xs:annotation>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name="rnaExperimentalForm" minOccurs="0" maxOccurs="unbounded">
<xs:annotation>
<xs:documentation>Describes the experimental form of RNA in a specific experiment.</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:sequence>
<xs:element name="experimentRef" type="refType">
<xs:annotation>
<xs:documentation>This experimental form has been used in the experiment referred to.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="rnaParticipantRef" type="refType">
<xs:annotation>
<xs:documentation>A reference to the experimental form of the participant.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="isOverexpressed" type="xs:boolean" minOccurs="0">
<xs:annotation>
<xs:documentation>True if the RNA has been overexpressed in the experiment.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="experimentalForm" type="cvType" minOccurs="0">
<xs:annotation>
<xs:documentation>The form of the participant in the interaction.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="experimentalRole" type="cvType" minOccurs="0">
<xs:annotation>
<xs:documentation>The role of the participant in the interaction.</xs:documentation>
</xs:annotation>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
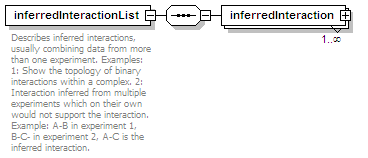
<xs:element name="inferredInteractionList" minOccurs="0">
<xs:annotation>
<xs:documentation>Describes inferred interactions, usually combining data from more than one experiment. Examples: 1: Show the topology of binary interactions within a complex. 2: Interaction inferred from multiple experiments which on their own would not support the interaction. Example: A-B in experiment 1, B-C- in experiment 2, A-C is the inferred interaction.</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:sequence>
<xs:element name="inferredInteraction" maxOccurs="unbounded">
<xs:complexType>
<xs:sequence>
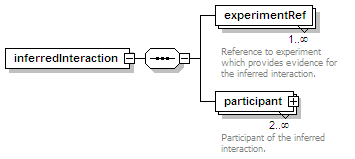
<xs:element name="experimentRef" type="refType" maxOccurs="unbounded">
<xs:annotation>
<xs:documentation>Reference to experiment which provides evidence for the inferred interaction.</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="participant" minOccurs="2" maxOccurs="unbounded">
<xs:annotation>
<xs:documentation>Participant of the inferred interaction.</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:choice>
<xs:element name="proteinParticipantRef" type="refType"/>
<xs:element name="proteinFeatureRef" type="refType"/>
<xs:element name="dnaParticipantRef" type="refType"/>
<xs:element name="dnaFeatureRef" type="refType"/>
<xs:element name="rnaParticipantRef" type="refType"/>
<xs:element name="rnaFeatureRef" type="refType"/>
<xs:element name="smallMoleculeParticipantRef" type="refType"/>
</xs:choice>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
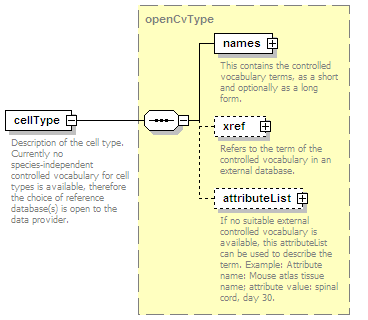
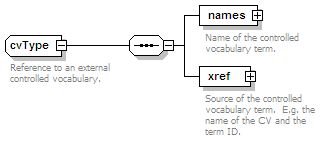
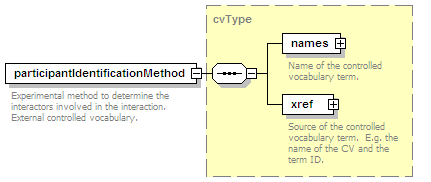
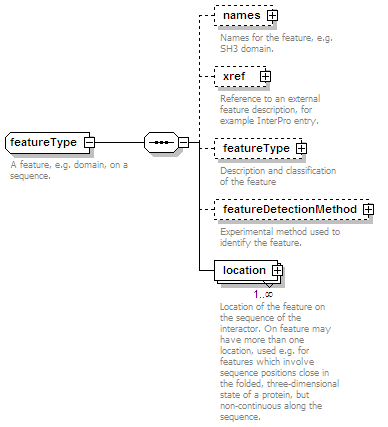
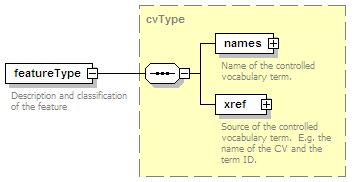
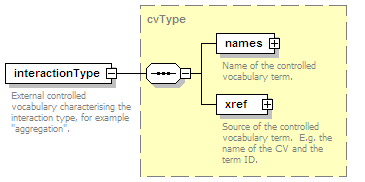
<xs:element name="interactionType" type="cvType" minOccurs="0" maxOccurs="unbounded">
<xs:annotation>
<xs:documentation>External controlled vocabulary characterising the interaction type, for example "aggregation".</xs:documentation>
</xs:annotation>
</xs:element>
<xs:element name="negative" type="xs:boolean" default="false" minOccurs="0">
<xs:annotation>
<xs:documentation>If true, this interaction has been shown NOT to occur under the described experimental conditions. Default false. If this optional attribute is missing, it is assumed to be set to false.</xs:documentation>
</xs:annotation>
</xs:element>
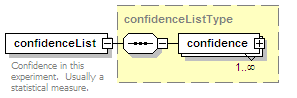
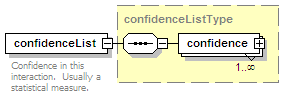
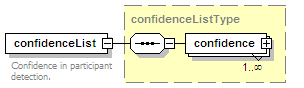
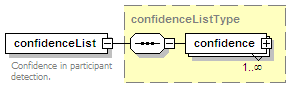
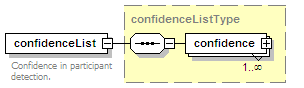
<xs:element name="confidenceList" type="confidenceListType" minOccurs="0">
<xs:annotation>
<xs:documentation>Confidence in this interaction. Usually a statistical measure.</xs:documentation>
</xs:annotation>
</xs:element>
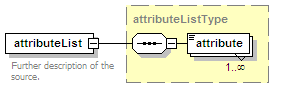
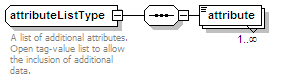
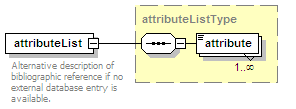
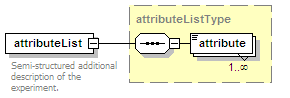
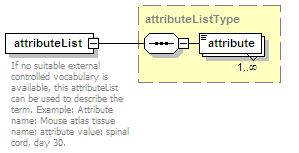
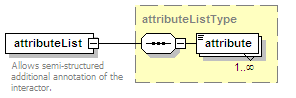
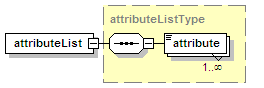
<xs:element name="attributeList" type="attributeListType" minOccurs="0">
<xs:annotation>
<xs:documentation>Semi-structured additional description of the data contained in the entry.</xs:documentation>
</xs:annotation>
</xs:element>
</xs:sequence>
</xs:complexType> |