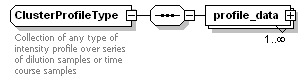
| content | complex |
| documentation |
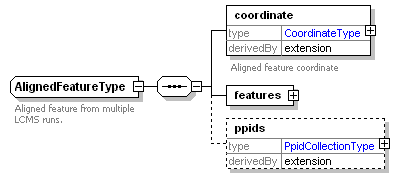
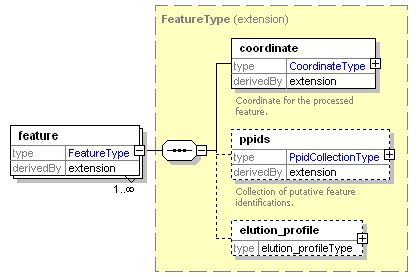
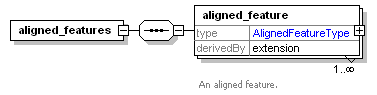
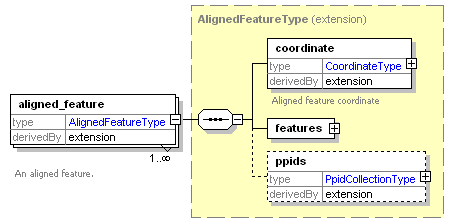
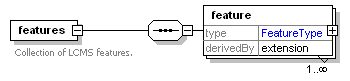
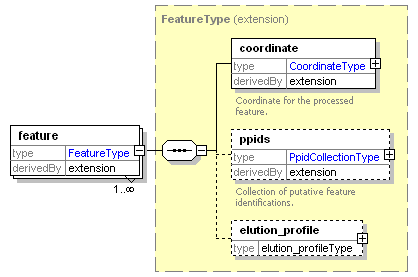
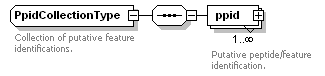
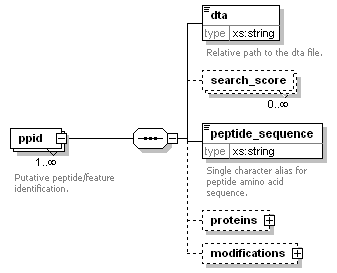
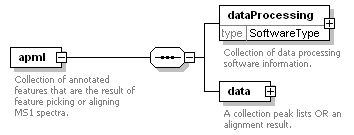
| Collection of annotated features that are the result of feature picking or aligning MS1 spectra. |
<xs:annotation>
<xs:documentation>Collection of annotated features that are the result of feature picking or aligning MS1 spectra.</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:sequence>
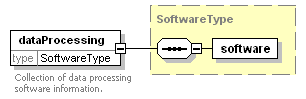
<xs:element name="dataProcessing" type="SoftwareType">
<xs:annotation>
<xs:documentation>Collection of data processing software information.</xs:documentation>
</xs:annotation>
</xs:element>
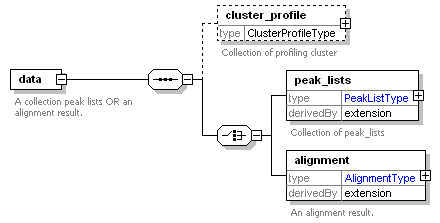
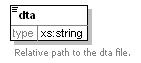
<xs:element name="data">
<xs:annotation>
<xs:documentation>A collection peak lists OR an alignment result.</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:sequence>
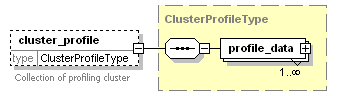
<xs:element name="cluster_profile" type="ClusterProfileType" minOccurs="0">
<xs:annotation>
<xs:documentation>Collection of profiling cluster</xs:documentation>
</xs:annotation>
</xs:element>
<xs:choice>
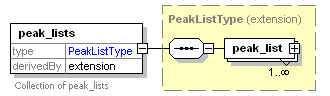
<xs:element name="peak_lists">
<xs:annotation>
<xs:documentation>Collection of peak_lists</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:complexContent>
<xs:extension base="PeakListType">
<xs:attribute name="count" type="xs:nonNegativeInteger" use="required"/>
</xs:extension>
</xs:complexContent>
</xs:complexType>
</xs:element>
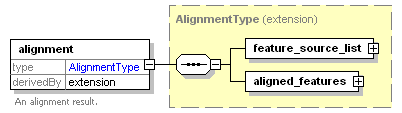
<xs:element name="alignment">
<xs:annotation>
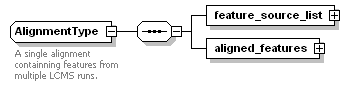
<xs:documentation>An alignment result.</xs:documentation>
</xs:annotation>
<xs:complexType>
<xs:complexContent>
<xs:extension base="AlignmentType"/>
</xs:complexContent>
</xs:complexType>
</xs:element>
</xs:choice>
</xs:sequence>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>